Abstract
Using dual-color fluorescence in situ hybridization (FISH) combined with two-dimensional (2D) image analysis, the locations of ABL and BCR genes in cell nuclei were studied. The center of nucleus-to-gene and mutual distances of ABL and BCR genes in interphase nuclei of nonstimulated and stimulated lymphocytes as well as in lymphocytes stimulated after irradiation were determined. We found that, after stimulation, the ABL and BCR genes move towards the membrane, their mutual distances increase, and the shortest distance between heterologous ABL and BCR genes increases. The distribution of the shortest distances between ABL and BCR genes in the G0 phase of lymphocytes corresponds to the theoretical distribution calculated by the Monte-Carlo simulation. Interestingly, the shortest ABL-BCR distances in G1 and S(G2 ) nuclei are greater in experiment as compared with theory. This result suggests the existence of a certain regularity in the gene arrangement in the G1 and S(G2 ) nuclei that keeps ABL and BCR genes at longer than random distances. On the other hand, in about 2% to 8% of lymphocytes, the ABL and BCR genes are very close to each other (the distance is less than ∼0.2 to 0.3 μm). For comparison, we studied another pair of genes, c-MYC and IgH, that are critical for the induction of t(8; 14) translocation that occurs in the Burkitt's lymphoma. We found that in about 8% of lymphocytes, c-MYC and IgH are very close to each other. Similar results were obtained for human fibroblasts. γ-Radiation leads to substantial changes in the chromatin structure of stimulated lymphocytes: ABL and BCR genes are shifted to the nuclear center, and mutual ABL-BCR distances become much shorter in the G1 and S(G2 ) nuclei. Therefore, we hypothesize that the changes of chromatin structure in the irradiated lymphocytes might increase the probability of a translocation during G1 and S(G2 ) stages of the cell cycle. The fact that the genes involved in the t(8; 14) translocation are also located close together in a certain fraction of cells substantiates the hypothesis that physical distance plays an important role in the processes leading to the translocations that are responsible for oncogenic transformation of cells.
CHROMOSOMAL abnormalities, including translocations, inversions, insertions, deletions, and numerical aberrations, are frequently detected in human cancer and particularly in leukemias.1,2 Many of these changes are not random: they have been found specific to a particular subtype of leukemia and often represent the only visible chromosomal alteration in the leukemic cells.2,3 The phenomenon of cytogenetic-clinicopathologic correlation implies that the genetic event resulting from a specific chromosomal rearrangement must be very important for the pathogenesis of leukemia.2-4
Translocations or exchanges of genetic material between chromosomes represent the best characterized cytogenetic abnormalities. The prototype of a translocation associated with leukemia is the t(9; 22)(q34; q11) that results in an abnormal chromosome 22 called the Philadelphia chromosome (Ph).4 This translocation involves an aberrant transfer of the c-abl protooncogene from chromosome 9 to the site adjoining the breakpoint cluster region (bcr) of the BCR gene on chromosome 22.2 The chimeric BCR-ABL gene created by the interchromosomal exchange produces a hybrid mRNA that translates the fusion protein.5,6 This phenomenon is fundamental in the pathogenesis of chronic myeloid leukemia (CML) and also in a subset of acute lymphoblastic leukemias and acute myelogenous leukemias.7-9 This chimeric gene is responsible for indolent clonal expansion, with an invariable progression to an aggressive phenotype terminated by blast crisis.7,10 The breakpoints of ABL and BCR genes vary widely in location; however, they always occur within introns, resulting in the fusion of specific ABL exons in the processed BCR-ABL mRNA. Breakpoints of the ABL gene occur in a large (200-kb) region in the 5′ portion of the gene, usually in the intron 1, whereas breakpoints in the BCR gene occur either between exons 2 and 3 (or 3 and 4; Mbcr) or between exons 1 and 2 (mbcr). Thus, two types of Mabl-bcr transcripts can be observed that join BCR exons 2 or 3 with ABL exons 2 through 11. This translocation is observed only in CML. The mbcr translocations result in fusion of the first exon of the BCR gene with ABL exons 2 through 11.2 10
Direct studies of atomic bomb survivors and patients treated by x-rays provide strong evidence that ionizing radiation induces both chronic myeloid leukemia and acute types of leukemia.11 The time trends are as follows: the death rate is already increasing at 2 years after the first exposure, peaking at 3 to 5 years, and declining to the control level more than 20 years after the exposure. From these facts, it seems that radiation induces the formation of BCR-ABL chimeric genes. However, the low frequency of such events makes the finding of direct evidence very difficult.12
The mechanism of ABL-BCR translocation is not known. It is possible that the distance between breakpoint regions in the replicating interphase nucleus influences the exchange of genetic material between the terminal q parts of chromosomes 9 and 22.
Only limited information about the arrangement of chromosomes and individual genes in the interphase nucleus exists. The nuclear architecture of the eukaryotic genome might be mediated by chromatin attachments to the nuclear matrix and nuclear membrane13,14 and by higher-order chromatin structures.15 Individual chromosomes have been shown to occupy distinct territories within mammalian nuclei.16-18 The results of optical space sectioning of polytene chromosomes of Drosophila larvae show that two apparently identical cells often have different chromosomes as nearest neighbors.19 Application of the fluorescence in situ hybridization (FISH) technique combined with improved optical microscopy and image processing systems have made it possible to visualize selected chromosomal domains in the interphase nuclei. Results obtained with probes of the constitutive heterochromatin of centromeric regions show their nonrandom distribution.20-22 Changes in the heterochromatin position of living neurons23 and a cell-cycle dependent clustering of human centromeres in the G1 phase that are dispersed as cells move into the S phase (observed by Bartholdi24 ) provide evidence that the centromeric heterochromatin might be specifically localized in certain cell types, whereas it is randomly distributed in others. Höfers et al25 suggest that these conclusions on localization of centromeric heterochromatin could be valid for any chromosome locus.
To obtain information about localization of abl and bcr regions in the interphase nuclei of human lymphocytes, we analyzed images of cell nuclei obtained semiautomatically with a high-resolution CCD camera using software that characterizes the positions of two FISH signals of different colors. We used an Mbcr probe modified with digoxigenin and an abl probe modified with biotin. The following measurements were taken: (1) the distances from the center of nucleus to genes ABL and BCR; (2) the distance between the two homologous green-stained ABL genes; (3) the distance between the two homologous red-stained BCR genes; and (4) the distance between the ABL and BCR genes.
Because of the high frequency of leukemias in individuals exposed to ionizing radiation, we performed similar measurements in lymphocytes irradiated before stimulation with 5 Gy of γ-rays to detect possible changes in the localization of ABL and BCR genes induced by the radiation.
To further support the hypothesis that a short distance between genes participating in translocation increases the probability of such an event, we investigated the distances between c-MYC and IgH loci in human lymphocytes and fibroblasts. The telomere 14q probe used in our experiments hybridizes to specific sequences at 14q32.3 that are located near the heavy-chain locus of immunoglobulin (IgH).26,27 Translocations that involve the IgH locus are very frequent in many B-cell tumors; this genetic lesion results in the deregulation of an oncogene as a consequence of the translocation. Activation of the c-MYC oncogene is a central event in Burkitt's lymphoma. A characteristic chromosomal translocation t(8; 14)(q24; q32), seen in 75% to 85% of patients, puts the c-MYC oncogene close to an IgH locus. Presumably, this rearrangement arises by random malfunctioning of the recombinase during maturation of B cells.27-29 Exchange of DNA between the two chromosomal positions implies that they are physically close to each other at the time this event occurs.30 Therefore, the occurence of a fraction of cells with c-MYC and IgH loci in close vicinity will substantiate the hypothesis that physical distance plays an important role in the processes leading to translocations responsible for oncogenic transformation.
MATERIALS AND METHODS
Cultivation and Irradiation of Lymphocytes
The lymphocytes were isolated from the whole heparinized blood (16 to 20 U/mL) of healthy individuals. Plasma containing lymphocytes was obtained after 2 hours of blood sedimentation at room temperature. It was divided into two 3-mL aliquots and irradiated by 0 and 5 Gy of 137Cs γ-rays (0.66 MeV; dose rate 3.6 Gy/min), respectively. Control and irradiated samples were diluted (1:3) with RPMI-1640 medium containing 0.1 mL of phytohaemagglutinin (PHA; Murex, Dartford, UK) per 5 mL of the suspension, 1% glucose, penicillin (100 U/mL), and streptomycin (100 μg/mL). The incubation was performed at 37°C for 50 hours. Colcemid was added 2 hours before the end of incubation. After hypotonic treatment (0.075 mol/L KCl), the nuclei were fixed in methanol/acetic acid (3:1) and the suspension was dropped on slides and allowed to dry.
Cell Cycle Analysis Using a Microscopic Method
The cell nuclei were classified as G0 , G1 , and S(G2 ) using a microscopic method.31 Two main characteristics were determined microscopically: the radius of cell nucleus and the number of doublets, ie, signals on both chromatids after DNA duplication. Small nuclei (radius <5 μm) were considered as nonstimulated in the G0 phase of the cell cycle.31 Large nuclei with only one signal of each of the ABL and BCR genes were grouped in the G1 phase; the nuclei with a double signal for at least one of these genes were grouped in the S(G2 ) phase. Nonstimulated lymphocytes were distinguished from other white blood cells by their large and round nucleus. Results were compared with flow cytometric data (G0+G1 , S, and G2 ) obtained at different periods after stimulation of both control and irradiated lymphocytes. Another criterion used was the number of centromeres31 that were visualized using a pancentromeric DNA probe.
Flow Cytometric Analysis
Lymphocytes were fixed in three successive baths of methanol/acetic acid mix (3:1) at −20°C for 15 minutes each. After washing in phosphate-buffered saline, the cells were resuspended in a buffer containing 50 μg/mL propidium iodide, 10 mmol/L Tris-HCl, pH 7.5, 5 mmol/L MgCl2 , and 10 μg/mL RNase A. Finally, the cells were analyzed on a Coulter Epics XL flow cytometer (Coulter, Hialeah, FL). In most cases, 2 × 104 cells were recorded per sample. DNA distribution was calculated using standard software.
DNA Probes, In Situ Hybridization, and Probe Detection
Mbcr-abl (digoxigenated bcr, biotinylated abl), digoxigenated c-myc, and biotinylated unique telomere 14q DNA probes were purchased from Oncor (Gaithersburg, MD), hybridized, and detected according to earlier published recommendations,32 with small modifications. The chromosomes were denatured at 70°C in 70% formamide with 2× SSC, pH 7, for 2 minutes; dehydrated in cold (−20°C) 70%, 80%, and 95% ethanol (2 minutes each); and then air dried.
The probe was prewarmed at 37°C for 5 minutes and vortexed. The Mbcr-abl DNA probe (10 μL) or a mixture of 10 μL telomere 14 and 7 μL of c-myc probes was applied to the denatured and dry slide, covered by a glass coverslip of 25 × 25 mm, sealed with rubber cement, and incubated overnight at 37°C in a humidified chamber in a perfect horizontal position.
Posthybridization washing was performed in 50% formamide with 2× SSC, pH 7, at 43°C for 15 minutes, followed by two 5-minute washes in 2× SSC containing 0.1% Tween 20, pH 7, at 37°C. Slides were then transferred to 1× PBD (Oncor) and incubated at room temperature for 3 minutes. Rhodamine-antidigoxigenin and fluorescein isothiocyanate (FITC)-avidin (60 μL) was applied to a slide and incubated for detection under a plastic coverslip at 37°C for 5 minutes. The washing was performed in three changes of 4× SSC containing 0.1% Tween 20, pH 7, at 43°C for 5 minutes. The temperature of the slides was equilibrated to room temperature in the 4× SSC containing 0.1% Tween 20, and the cells were contrastained with 4′,6-diamidine-2′-phenylindole (DAPI; 0.02 μg/mL) in the Vectashield (Vector, Burlingame, CA).
Distribution of Lymphocytes in Different Phases of the Cell Cycle as a Function of the Time of Stimulation Determined From Flow Cytometric and Microscopic Data
| Radiation Dose (Gy) . | Time of Stimulation (h) . | No. of Cells (%) . | |||
|---|---|---|---|---|---|
| . | . | G0 . | G1 . | S . | G2 . |
| 0 | 83.0 ± 5.4 | 14.0 ± 2.9 | 3.0 ± 2.9 | 0 ± 2.5 | |
| 0 | 20 | 56.0 ± 6.3 | 28.2 ± 2.8 | 14.5 ± 3.8 | 1.3 ± 2.5 |
| 30 | 17.0 ± 6.9 | 65.1 ± 4.4 | 13.2 ± 4.1 | 4.7 ± 2.8 | |
| 40 | 78.7 ± 6.3* | 15.8 ± 3.6 | 5.5 ± 2.7 | ||
| 50 | 25.0 ± 2.6 | 54.7 ± 4.2 | 10.6 ± 3.7 | 9.7 ± 3.1 | |
| 20 | 80.9 ± 6.2* | 12.5 ± 3.4 | 6.5 ± 2.8 | ||
| 5 | 30 | 30.0 ± 2.7 | 43.4 ± 3.0 | 17.6 ± 3.1 | 8.0 ± 2.6 |
| 40 | 76.2 ± 6.9* | 13.7 ± 3.8 | 10.1 ± 3.1 | ||
| 50 | 26.0 ± 2.5 | 51.7 ± 5.0 | 12.0 ± 4.1 | 10.3 ± 3.4 | |
| Radiation Dose (Gy) . | Time of Stimulation (h) . | No. of Cells (%) . | |||
|---|---|---|---|---|---|
| . | . | G0 . | G1 . | S . | G2 . |
| 0 | 83.0 ± 5.4 | 14.0 ± 2.9 | 3.0 ± 2.9 | 0 ± 2.5 | |
| 0 | 20 | 56.0 ± 6.3 | 28.2 ± 2.8 | 14.5 ± 3.8 | 1.3 ± 2.5 |
| 30 | 17.0 ± 6.9 | 65.1 ± 4.4 | 13.2 ± 4.1 | 4.7 ± 2.8 | |
| 40 | 78.7 ± 6.3* | 15.8 ± 3.6 | 5.5 ± 2.7 | ||
| 50 | 25.0 ± 2.6 | 54.7 ± 4.2 | 10.6 ± 3.7 | 9.7 ± 3.1 | |
| 20 | 80.9 ± 6.2* | 12.5 ± 3.4 | 6.5 ± 2.8 | ||
| 5 | 30 | 30.0 ± 2.7 | 43.4 ± 3.0 | 17.6 ± 3.1 | 8.0 ± 2.6 |
| 40 | 76.2 ± 6.9* | 13.7 ± 3.8 | 10.1 ± 3.1 | ||
| 50 | 26.0 ± 2.5 | 51.7 ± 5.0 | 12.0 ± 4.1 | 10.3 ± 3.4 | |
The cell cycle phases were determined by flow cytometry (G0 + G1 , S, and G2 ) and according to the nuclear radius (R)31 (R < 5 μm for G0 , R > 5 μm for G1 + S + G2 ).
Microscopic analysis was not performed.
Data Acquisition and Statistical Analysis
The green and red fluorescent signals for abl and bcr genes were detected on a blue background. Digital images were generated using a single-chip cooled color camera (Hamamatsu C5310; Hamamatsu Photonics K.K., Shimokanzo, Japan). The camera was attached to a Jenalumar epifluorescence microscope (Carl Zeiss GmbH, Jena, Germany) equipped with dual-band and triple-band filters (AHF, Tübingen, Germany) for FITC-PI and FITC-PI-DAPI, respectively. The images were digitized (MuTech Image/VGA Plus frame-grabber; MuTech Corp, Woburn, MA) with 24-bit resolution (8 bits per color) and processed using a Pentium 100 MHz computer (AutoCont s.r.o., Brno, Czech Republic).
A special software, FISH 1.1 (M. Kozubek, Institute of Biophysics, Academy of Sciences, Brno, Czech Republic), was developed for the automated analysis of the FISH-painted nuclei.33 FISH 1.1 runs under the MS-Windows environment and contains a number of filters and procedures for image analysis.
The nuclei were detected using simple thresholding in blue color. The optimal threshold was found by an automatic histogram analysis. The green and red signals inside nuclei were detected by means of a modified watershed algorithm.33 34
To detect exactly two green (red) signals for ABL (BCR) genes, the following steps were applied. (1) The nucleus was excluded from the final data set if no or just one green (red) signal was found. (2) The nucleus was excluded if the intensity of the third highest (green or red) signal found on the nucleus was stronger than two thirds of the intensity of the second highest signal. (3) The nucleus was accepted otherwise, taking into account only the two highest signals.
The coordinates of the signals, the distance of the signals from the center of the nucleus (in percentage of local radius), their intensity, and their area were computed. To confirm the results of the analysis, the signals were visualized on a screen. After the analysis of each image, text files were generated with information about the image, each nucleus on the image, and each signal on a given nucleus. An appropriate format of the text file was chosen for the import of the whole data set into SigmaPlot (Jandel Scientific, San Rafael, CA). Two-dimensional (2D) projections of the distances between the signals and additional characteristics were calculated using the Transform option of SigmaPlot. All distances between fluorescent signals in the interphase nuclei of lymphocytes were determined as projections. The significance of the differences between various distributions was calculated using the Student's t-test option of SigmaPlot at P = .05.
The lymphocytes increased in diameter after stimulation [from ∼10 μm in the G0 phase up to 16 μm in the S(G2 ) phase]. Therefore, all distances are expressed as the percentage of nuclear radius.
Calculation of Theoretical Distributions
The following notation will be used throughout the remainder of this paper: (1) AA, distances between homologous ABL-ABL genes; (2) BB, distances between homologous BCR-BCR genes; (3) ABm , minimum of four distances ABL-BCR in nucleus; (4) CA, distances between nuclear weight center and ABL gene in terms of percentage of local radius (local radius is the distance between the nuclear center and the membrane in the direction of the ABL [BCR] gene); (5) CB, distances between nuclear weight center and BCR gene. The average values of the above-mentioned distances will be AA, BB, CA, CB, and ABm. In the case of dual-color probes for telomere 14q and c-MYC, the same notation will be used with the letters T and M, ie, TT, MM, TMm , CT, and CM.
It will be shown later that in all phases of the cell cycle of lymphocytes the distribution of CA and CB distances does not correspond to random location of the ABL and BCR genes in a sphere. The average distance of a point, randomly placed in a sphere, from the center of the sphere in the 2D projection is 58.8% of radius, which is much more than CA and CB (see the Results). It means that the genes are situated preferentially at certain distances from the center. Reconstruction of the three-dimensional distribution of CA or CB distances is possible under the condition that the nuclei are rotationally symmetrical, ie, that the distribution of genes in spherical concentric layers is random (although the genes can be situated preferentially at certain layers). This approach will be called the model of rotational symmetry (RS model). Calculation of the distributions in the RS model is quite simple. Four points (simulating ABL and BCR genes) are placed in a sphere using weighted random number generator with different weights at different layers. For each quadruple of points, the values of AA, BB, ABm , CA, and CB are calculated. The procedure is repeated about 105 times, and distributions of all distances are generated. The probabilities of gene appearance in different layers (weights of random number generator) are adjusted in such a way that the theoretical distributions of CA and CB distances in projection reproduce the experimental histograms.
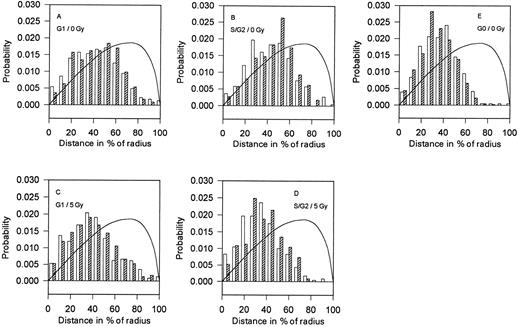
Distances between the nuclear center and BCR (ABL) genes (CB and CA). Distribution of the 2D projections of the distances between the BCR (□) or ABL (▨) genes and the center of nucleus (CB and CA) of the control (A, B, and E) and the γ-irradiated (5 Gy; C and D) human lymphocytes. The mean values and standard deviations are given in Table 2. The corresponding theoretical distributions of the 2D projections of the distances between the center of the sphere and the point randomly placed inside the sphere are shown by the solid line.
Distances between the nuclear center and BCR (ABL) genes (CB and CA). Distribution of the 2D projections of the distances between the BCR (□) or ABL (▨) genes and the center of nucleus (CB and CA) of the control (A, B, and E) and the γ-irradiated (5 Gy; C and D) human lymphocytes. The mean values and standard deviations are given in Table 2. The corresponding theoretical distributions of the 2D projections of the distances between the center of the sphere and the point randomly placed inside the sphere are shown by the solid line.
Using the RS model, the distributions of AA, BB, and ABm distances based on experimental CA and CB histograms were calculated and compared with the experimental AA, BB, and ABm histograms.
RESULTS
The center-to-gene distances as well as the distances between genes in nuclei were measured in the intact and irradiated lymphocytes both immediately after the addition of PHA and after 50 hours of incubation. All experiments were repeated three times; for each iteration, 200 cell nuclei were analyzed.
After 50 hours of incubation, the lymphocytes are mostly stimulated. The proportion of cells in the G0 , G1 , and S(G2 ) stages was approximately 30%, 40% to 50%, and 20% to 30%, respectively, in both the control and irradiated samples. Flow cytometric and microscopic data that express the distribution of lymphocytes in the different stages of the cell cycle as a function of the time of stimulation are shown in Table 1.
Center-to-Gene Distributions of ABL and BCR Genes (CA and CB Distributions)
Nonirradiated controls.The center-to-gene distributions of the ABL and BCR genes (CA and CB distributions) are shown in Fig 1 A and B. The mean values of these distances (CA and CB) are significantly lower for cells in the G0 phase compared with cells in the G1 and S(G2 ) phases (Table 2). The shape of the distributions of nuclear center-to-gene distances changes from narrow for cells in the G0 phase to broad in the G1 phase and again to narrow one in the S(G2 ) phase (Fig 1A, B, and E). The distances CB are significantly longer than the distances CA for cells in the G0 phase; they are equal in the G1 phase and shorter in the S(G2 ) phase (Table 2). Thus, for cells in the G0 phase, chromosome 22 is more distant from the nuclear center than chromosome 9, whereas chromosome 22 is closer to the nuclear center for cells in the S(G2 ) phase.
The Average Values and Standard Deviations of the AA, BB, ABm , CA, and CB Distributions in Interphase Nuclei of Control and Irradiated Lymphocytes
| Phase of Cell Cycle . | Irradiation (Gy) . | AA (% of R)
. | BB (% of R)
. | AB m (% of R)*
. | CA (% of R)
. | CB (% of R)
. |
|---|---|---|---|---|---|---|
| G0 | 0 | 57.3 ± 1.5 (55.8)† | 53.4 ± 1.3 (52.2) | 24.0 ± 0.9 (25.4) | 34.9 ± 0.7 | 37.4 ± 0.7 |
| G1 | 0 | 63.0 ± 1.8 (59.3) | 57.9 ± 1.7 (55.2) | 34.5 ± 1.1 (30.2)‡ | 43.5 ± 0.8 | 42.9 ± 0.9 |
| S(G2 ) | 0 | 61.9 ± 1.8 (60.9) | 59.4 ± 1.5 (58.7) | 36.0 ± 1.0 (31.7)‡ | 45.6 ± 0.7 | 43.3 ± 0.7 |
| G1 | 5 | 60.2 ± 1.4 (54.2) | 59.8 ± 1.7 (55.5) | 26.6 ± 1.0 (28.6) | 39.6 ± 0.8 | 40.0 ± 0.9 |
| S(G2 ) | 5 | 52.1 ± 1.2 (50.9) | 49.7 ± 1.4 (47.1) | 28.4 ± 0.9 (27.4) | 37.3 ± 0.7 | 35.1 ± 0.7 |
| G1 (5 Gy/0 Gy) | 0.957 ± 0.049 | 1.033 ± 0.058 | 0.772 ± 0.052ρ | 0.911 ± 0.0352-155 | 0.933 ± 0.0382-155 | |
| S(G2 ) (5 Gy/0 Gy) | 0.842 ± 0.044ρ | 0.837 ± 0.044ρ | 0.789 ± 0.047ρ | 0.818 ± 0.027ρ | 0.811 ± 0.031ρ | |
| 0 Gy (G1 /S) | 1.017 ± 0.058 | 0.975 ± 0.052 | 0.957 ± 0.056 | 0.954 ± 0.031 | 0.990 ± 0.037 | |
| 5 Gy (G1 /S) | 1.156 ± 0.053ρ | 1.204 ± 0.0672-155 | 0.936 ± 0.066 | 1.062 ± 0.040 | 1.139 ± 0.048 | |
| Phase of Cell Cycle . | Irradiation (Gy) . | AA (% of R)
. | BB (% of R)
. | AB m (% of R)*
. | CA (% of R)
. | CB (% of R)
. |
|---|---|---|---|---|---|---|
| G0 | 0 | 57.3 ± 1.5 (55.8)† | 53.4 ± 1.3 (52.2) | 24.0 ± 0.9 (25.4) | 34.9 ± 0.7 | 37.4 ± 0.7 |
| G1 | 0 | 63.0 ± 1.8 (59.3) | 57.9 ± 1.7 (55.2) | 34.5 ± 1.1 (30.2)‡ | 43.5 ± 0.8 | 42.9 ± 0.9 |
| S(G2 ) | 0 | 61.9 ± 1.8 (60.9) | 59.4 ± 1.5 (58.7) | 36.0 ± 1.0 (31.7)‡ | 45.6 ± 0.7 | 43.3 ± 0.7 |
| G1 | 5 | 60.2 ± 1.4 (54.2) | 59.8 ± 1.7 (55.5) | 26.6 ± 1.0 (28.6) | 39.6 ± 0.8 | 40.0 ± 0.9 |
| S(G2 ) | 5 | 52.1 ± 1.2 (50.9) | 49.7 ± 1.4 (47.1) | 28.4 ± 0.9 (27.4) | 37.3 ± 0.7 | 35.1 ± 0.7 |
| G1 (5 Gy/0 Gy) | 0.957 ± 0.049 | 1.033 ± 0.058 | 0.772 ± 0.052ρ | 0.911 ± 0.0352-155 | 0.933 ± 0.0382-155 | |
| S(G2 ) (5 Gy/0 Gy) | 0.842 ± 0.044ρ | 0.837 ± 0.044ρ | 0.789 ± 0.047ρ | 0.818 ± 0.027ρ | 0.811 ± 0.031ρ | |
| 0 Gy (G1 /S) | 1.017 ± 0.058 | 0.975 ± 0.052 | 0.957 ± 0.056 | 0.954 ± 0.031 | 0.990 ± 0.037 | |
| 5 Gy (G1 /S) | 1.156 ± 0.053ρ | 1.204 ± 0.0672-155 | 0.936 ± 0.066 | 1.062 ± 0.040 | 1.139 ± 0.048 | |
The distances are shown as a percentage of the nuclear radius (R).
Abbreviations: AA, average distance ABL-ABL;BB, average distance BCR-BCR;ABm , average minimum distance ABL-BCR;CA, average distance center of nucleus-to-ABL;CB, average distance center of nucleus-to-BCR.
Minimum of 4 distances between 2D projections of ABL and BCR genes in a nucleus is taken.
Theoretical values (in parentheses); the standard errors of the theoretical values (derived from the standard errors of the average center-to-gene distances) are less than that of experimental ones.
Theoretical value is significantly different from the experimental one (P < .05).
ρ Significantly different from 1 (P < .005).
Significantly different from 1 (P < .05).
After γ-irradiation.The ABL and BCR genes are located closer to the nuclear center in both the G1 and S(G2 ) lymphocytes irradiated with 5Gy of γ-rays (Fig 1C and D). Compared with control samples, the distributions have smaller distances. Changes in the S(G2 ) phase are more pronounced compared with cells in the G1 phase. The difference between the center-to-gene average distances of ABL and BCR genes (CA and CB) is not statistically significant for cells in the G1 phase; at the same time, for cells in the S(G2 ) phase, the center-to-gene distances for BCR genes are shorter than for ABL genes (CB<CA). The results clearly show that, after irradiation, ABL and BCR genes are situated in close proximity to the nuclear center compared with the nonirradiated control.
Comparison of theoretical, random distributions of genes in a sphere with our experimental data strongly suggests that the location of both ABL and BCR genes is not random for all stimulated, nonstimulated, irradiated, and nonirradiated lymphocytes (Fig 1A through E).
Distance Between Homologous ABL and BCR Genes (AA and BB Distances)
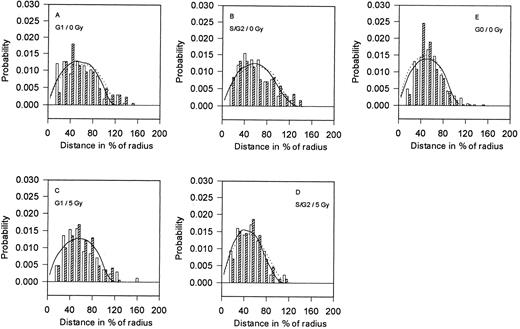
Nonirradiated controls.The distances between the homologous abl and bcr regions were determined for both irradiated and nonirradiated lymphocytes in the G0 , G1 , and S(G2 ) phases of the cell cycle (Fig 2A through E). The average distance between ABL(BCR) genes on homologous chromosomes in the G0 phase is significantly shorter than the same distance in the G1 or S(G2 ) phases (Table 2). Generally, in all of these cells, homologous BCR genes are closer together than homologous ABL genes (BB<AA). The differences in the distances of these genes in the G1 and S(G2 ) cell nuclei are not significant. Theoretical distributions of distances between homologous ABL and BCR genes calculated according to the RS model correspond quite well to the experimental ones (Fig 2A, B, and E); the only exceptions are distances between 30% to 50% of the nuclear radius that are more frequent than the theoretical distances.
Distances between homologous BCR and ABL genes (BB and AA). Distributions of 2D-projections of the distances between homologous BCR genes (□) and ABL genes (▨) in the control (A, B, and E) and in the γ-irradiated (5 Gy; C and D) human lymphocytes. The mean values and standard deviations are given in Table 2. The full (dotted) curves represent the corresponding theoretical distributions calculated by Monte-Carlo simulation using experimentally determined center-to-gene distributions from Fig 1 (according to the RS model).
Distances between homologous BCR and ABL genes (BB and AA). Distributions of 2D-projections of the distances between homologous BCR genes (□) and ABL genes (▨) in the control (A, B, and E) and in the γ-irradiated (5 Gy; C and D) human lymphocytes. The mean values and standard deviations are given in Table 2. The full (dotted) curves represent the corresponding theoretical distributions calculated by Monte-Carlo simulation using experimentally determined center-to-gene distributions from Fig 1 (according to the RS model).
After γ-irradiation.In irradiated nuclei, the distances between homologous ABL as well as BCR genes in the S(G2 ) stage of the cell cycle are substantially shorter than in nonirradiated nuclei (P < .05 for BCR and P < .02 for ABL regions; Fig 2C and D). These distances are not significantly different between irradiated and nonirradiated cells in the G1 phase (Table 2); however, in the irradiated nuclei, BCR genes are closer to each other.
Distance Between ABL and BCR Genes (ABm Distances)
Nonirradiated controls.The distances between each ABL gene (green) to one or another BCR gene (red) were determined. The minimum value of the four measured distances per cell nucleus was determined (ABm ). The distances ABm for each stage of the cell cycle are plotted as normed histograms in Fig 3A, B, and E. The distributions generated by Monte-Carlo calculations according to the RS model are shown by curves in Fig 3A, B, and E. In both G1 and S(G2 ) cell nuclei, the theoretical ABm distances between nonhomologous genes are significantly shorter (P < .05) than experimental ABm distances (Fig 3A and B and Table 2). No significant difference in the experimental distributions of ABm distances between cells in the G1 and S(G2 ) phases of the cell cycle exists. In the G0 phase, the detected distances between nonhomologous genes (Fig 3E) correspond to the theoretical prediction (solid line in Fig 3E and Table 2).
Minimum distances between nonhomologous BCR and ABL genes (ABm ). Distributions of 2D projections of the minimum distances between the BCR and ABL genes in the control (A, B, and E) and in the γ-irradiated (5 Gy; C and D) human lymphocytes. The mean values and standard deviations are given in Table 2. The curves represent the theoretical distributions calculated by Monte-Carlo simulation using experimentally determined center-to-gene distributions from Fig 1 (according to the RS model).
Minimum distances between nonhomologous BCR and ABL genes (ABm ). Distributions of 2D projections of the minimum distances between the BCR and ABL genes in the control (A, B, and E) and in the γ-irradiated (5 Gy; C and D) human lymphocytes. The mean values and standard deviations are given in Table 2. The curves represent the theoretical distributions calculated by Monte-Carlo simulation using experimentally determined center-to-gene distributions from Fig 1 (according to the RS model).
After γ-irradiation.In lymphocytes irradiated with 5 Gy of γ-rays, the average minimum distance between ABL and BCR genes is substantially shorter than in the nonirradiated lymphocytes (Fig 3C and D and Table 2). This distance (ABm ) decreases from 34.5% to 26.6% of the cellular radius for G1 cells and from 36.0% to 28.4% for the S(G2 ) cells (the differences are significant at P < .01). The percentage of cells in which one of the green signals (ABL) is located in very close proximity to one of the red (BCR) signals (≤0.2 to 0.3 μm) increases (Fig 3) from 2.0% ± 1.0% to 4.1% ± 1.8% in the G1 phase and from 1.5% ± 1.1% to 5.9% ± 2.2% in the S(G2 ) phase. Notably, the percentages for irradiated cells are substantially higher than the theoretically predicted ones (Fig 3C and D, the first column). Differences between irradiated and nonirradiated lymphocytes are not statistically significant. However, an increased number of lymphocytes with ABL and BCR genes located close together was repeatedly observed after irradiation; the same phenomenon was also observed in the fibroblasts in which the number of such nuclei was higher than predicted theoretically even in the nonirradiated cells (results not shown).
DISCUSSION
Using dual-color FISH combined with 2D image analysis, we measured the 2D projections of center of nucleus-to-gene distances for ABL and BCR genes and mutual distances between both homologous and heterologous ABL and BCR genes in interphase lymphocytes. We found that, after the stimulation of lymphocytes, the ABL and BCR genes move towards the membrane, their mutual distances increase, and the minimum distance between heterologous ABL and BCR genes increases. The experimental distribution of the shortest distances between ABL and BCR genes (ABm distribution) in the G0 phase of lymphocytes corresponds to the theoretical ABm distribution calculated according to the RS model (see the Materials and Methods). On the other hand, experimentally measured ABm distances are greater than ABm distances determined theoretically (using RS model) for cells in the G1 and S(G2 ) phases, giving evidence of a special type of nonrandom arrangement. The transition of lymphocytes from the G0 phase to the G1 and S(G2 ) phases involves rearrangement of chromatin manifested by prolongation of minimal distances between ABL and BCR genes that exceeds the calculated random distances. This nonrandom arrangement of ABL and BCR genes is also evidenced by the more frequent (as compared with the theoretical calculation) occurrence of cells with two homologous genes at a distance of 30% to 50% of the nuclear radius (Fig 2A and B). Nonrandom spatial distribution of ABL, BCR, and chimeric BCR-ABL genes was also observed in bone marrow cells of CML patients.35
Ionizing radiation partially inhibits the changes in ABL and BCR localization induced in the lymphocytes by stimulation with PHA. This effect is most pronounced during the S(G2 ) phase. In these irradiated lymphocytes, ABL and BCR genes are near the center of nucleus, the distances between both homologous and heterologous genes are shorter, and the minimum distance between heterologous genes is substantially shorter.
Ionizing radiation induces a wide spectrum of molecular lesions in DNA, including single-strand breaks, double-strand breaks, base damage, and DNA-protein cross-links.36,37 The majority of DNA breaks are repaired within 10 minutes after irradiation.38 However, the repair process of double-strand breaks involves mistakes that lead to chromosomal aberrations.36,38-40 Unrepaired or misrepaired double-strand breaks or chromosomal aberrations might be one possible cause of chromatine changes in stimulated lymphocytes. In addition, a loss of supercoiling ability was observed in irradiated cells.41 42 The investigators hypothesize that the loss of superhelical density could be attributed to the irreparable lesions induced by radiation. It is possible that the chromatine changes in irradiated lymphocytes observed in our experiments could be related to a lower superhelical density of chromatin resulting from irradiation.
The distribution of ABL-BCR distances in the irradiated cells corresponds quite well to the RS model. This result means that the nonrandom structural arrangement of ABL-BCR genes in the stimulated cells changed after irradiation. In a fraction of irradiated cell nuclei (about 5%), the ABL and BCR genes are very close to each other (<0.2 to 0.3 μm). This fraction is substantially larger than the fraction theoretically predicted from random superposition of signals in the 2D projection. The reason for this stickiness of ABL and BCR regions is unknown, but one possibility could be an association based on homology of DNA in certain regions of these genes.30 It is tempting to speculate that physical distance and translocation frequency between specific sequences are correlated. Direct evidence of the induction of BCR-ABL fusion by a high dose of X-irradiation in HL60 cells in vitro was reported by Ito.12
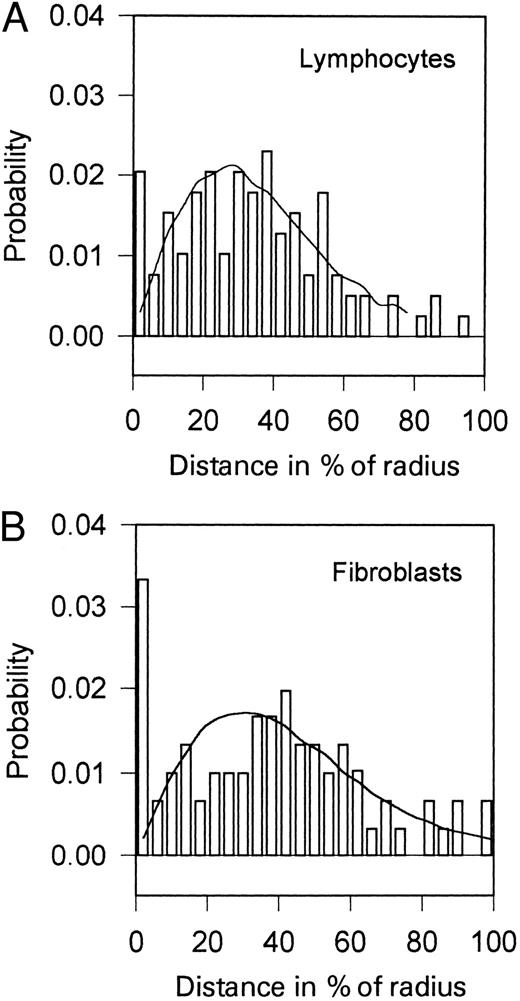
To further substantiate the hypothesis that physical distance of specific sequences increases the translocation frequency, we investigated the distance distributions of the c-MYC gene and unique telomere 14q sequences situated in close proximity of the IgH gene. Both of these locuses are involved in the translocation t(8; 14) (q24.1; q32.3) in Burkitt's lymphoma. It is supposed that the t(8; 14) translocation arises through a malfunction of the recombinases that rearrange the IgH gene during maturation of B cells.27-29 The results of our investigations using lymphocytes and fibroblasts are shown in Fig 4. Distances between heterologous c-MYC and telomere 14q sequences are distributed according to the RS model, with the exception of those cases in which the genes are very close to each other (<0.2 to 0.3 μm). The number of nuclei with c-MYC and telomere 14q sequences that are close to each other is many times higher than the number resulting from theoretical distribution. This difference is significant in both cell types (lymphocytes and fibroblasts), which supports the hypothesis that the association of these genes is related to some mechanism that is cell-type nonspecific. Homology of DNA sequences might serve as a basis of one of such mechanisms.
Distributions of the distances between c-MYC and unique telomere 14q sequences (TMm ; lying adjacent to the Ig heavy chain locus) in human lymphocytes incubated for 50 hours with PHA (A) and in human fibroblasts (B). The curves represent the theoretical distributions calculated by Monte-Carlo simulation using experimentally determined center-to-gene distributions (not shown). Human fibroblasts (VF-10) were grown in RPMI medium supplemented with 10% fetal calf serum, 0.5% L-glutamine, 1% glucose, and antibiotics at 37°C to the confluent state.
Distributions of the distances between c-MYC and unique telomere 14q sequences (TMm ; lying adjacent to the Ig heavy chain locus) in human lymphocytes incubated for 50 hours with PHA (A) and in human fibroblasts (B). The curves represent the theoretical distributions calculated by Monte-Carlo simulation using experimentally determined center-to-gene distributions (not shown). Human fibroblasts (VF-10) were grown in RPMI medium supplemented with 10% fetal calf serum, 0.5% L-glutamine, 1% glucose, and antibiotics at 37°C to the confluent state.
Exchange of DNA between two chromosomal positions implies that they are physically close together at the time this event occurs.30 Consequently, the frequency of translocations in cells should increase if the physical distance between critical genes is reduced. We found substantially reduced physical distance between ABL and BCR genes in irradiated human lymphocytes in the G1 and S(G2 ) stages after stimulation. We also found that the genes involved in the t(8; 14) translocation between c-MYC and IgH loci are frequently close to each other already in the control (nonirradiated) lymphocytes and fibroblasts. The proximity of c-MYC to IgH loci increases the accessibility of some of their sequences to the recombinase responsible for assemblage of the Ig gene during B-cell development. Through mistakes in this gene rearrangement mechanism, the translocation between c-MYC and IgH loci can appear in B cells. The translocations are manifested as Burkitt's lymphoma. The mechanism of Ig genes rearrangement is limited only to lymphoid cells; therefore, the physical proximity of c-MYC to IgH loci in fibroblasts cannot lead to their translocation.
We conclude that the locations of genes in the interphase nucleus are not fully random. Their organization can be characterized by the following principles. (1) The genes can be localized most commonly at a specific range of distances between the nuclear center and membrane (in a spherical layer). This type of nonrandom organization of genes in nuclei makes it possible to describe the structure by the RS model (see the Materials and Methods), which gives in many cases reasonably good results. (2) Two homologous or heterologous genes inside the layer can be located at preferential mutual positions. This principle is manifested by the discrepancy between experimentally observed distributions of gene-to-gene distances and the corresponding theoretical distribution calculated on the basis of RS model. (3) Two heterologous genes can be associated at very short distances (stickiness of genes). For homologous genes, this phenomenon is observed in S(G2 ) cells.
These principles are valid for the organization of cell nucleus in dependence on other factors such as the type of the cell, the phase of the cell cycle, the state of the cellular environment, and, of course, the characteristics of the genes themselves.
Supported by the Grant Agency of the Czech Republic (Grant No. 202/96/1718) and by the Grant Agency of the Ministry of Health of the Czech Republic (Grant No. 2636-2).
Address reprint requests to Dr Stanislav Kozubek, Institute of Biophysics, Academy of Sciences, Královopolská 135, 612 65 Brno, Czech Republic.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal