Abstract
Lutheran glycoprotein (Lu gp) has five predicted immunoglobulin superfamily (IgSF ) domains. K562 cells were transfected with Lu cDNA and tested by flow cytometry with monoclonal antibodies and Lu blood group antisera. The results confirmed the identity of Lu cDNA. Deletion mutants lacking the regions encoding one or more IgSF domains were made by inverse polymerase chain reaction (PCR), expressed in K562 cells, and tested with the same antibodies. The Lub and Lu5 antigens and the epitope recognized by monoclonal antibody BRIC 224 were mapped to the first, N-terminal, IgSF domain. Lu4 and Lu8 were mapped to domain 2; Lu20 to domain 3; Lu7 and BRIC 221 epitope to domain 4, and Lu13 and Aub to domain 5. The organization of the LU gene was determined. The region encoding the open reading frame is arranged in 15 exons extending over ≈11 kb on chromosome 19q13.2. The Lua/Lub and Aua/Aub blood group polymorphisms were studied using genomic DNA from typed blood donors. The Lua mutation is a base change in exon 3 (G252 to A) encoding an Arg77 (Lub) to His (Lua) change on the CFG face of domain 1. The Aua/Aub polymorphism is an A1637 to G substitution in exon 12 encoding a Thr539 (Aua) to Ala (Aub) change on the G strand of domain 5.
THE LUTHERAN (LU) BLOOD group system, which consists of 18 antigens, includes polymorphisms comprising four pairs of allelic antigens: Lua (Lu1) and Lub (Lu2); Lu6 and Lu9; Lu8 and Lu14; Aua (Lu18) and Aub (Lu19).1 The remainder are antigens of very high frequency, included in the Lu system because they are absent from red blood cells of the Lunull phenotype. Most of the Lu system antigens have been shown, by immunoblotting, to be located on the Mr 85,000 Lu glycoprotein (gp) and its Mr 78,000 isoform.2,3 Molecular characterization of the Lu blood group antigens has been hampered by the fact that it has not been possible to detect Lu mRNA in peripheral blood reticulocytes. Prior to this study, the organization of the LU gene, which is located on chromosome 19 q13.2,4 5 was unknown.
We have previously isolated a cDNA clone for the Lu gp from a placental cDNA library. The clone encodes a predicted protein that is a member of the immunoglobulin superfamily (IgSF ) of molecules with five extracellular, Ig-like domains: from the N-terminus 2 V-set and 3 C2-set domains (Fig 1).4 The predicted protein has a hydrophobic, transmembrane domain and a cytoplasmic domain of some 59 amino acid residues. An homologous cDNA clone5 (named B-CAM), which differs from Lu gp in that it lacks the majority of the cytoplasmic domain, has been shown to arise from alternate splicing of an immature LU gene transcript.6 The predominant isoform of Lu gp in most normal tissues is an Mr 85,000 glycoprotein corresponding to the full-length Lu cDNA, although in red blood cell membranes, the minor, Mr 78,000 spliceoform (presumed to be encoded by the B-CAM mRNA sequence) can also be identified.2,4,7 Lu gp has five potential N-glycosylation sites, with one site on the third Ig-like domain and four sites on the fourth domain.4 The function of Lu gp is unknown, although by analogy with other IgSF members and those with the domain structure V-V-C2-C2-C2, it would be predicted to have an adhesion and/or receptor function.
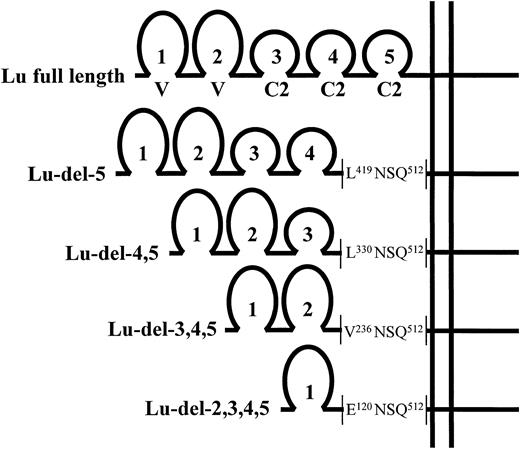
The IgSF domain structure of the expressed full-length and domain-deleted Lu cDNA constructs. The predicted amino acid sequence of the domain-deletion mutants at the point of the deletion, proximal to the predicted transmembrane domain, is shown.
The IgSF domain structure of the expressed full-length and domain-deleted Lu cDNA constructs. The predicted amino acid sequence of the domain-deletion mutants at the point of the deletion, proximal to the predicted transmembrane domain, is shown.
In this study, we have confirmed the identity of the Lu cDNA clone by eukaryotic expression in K562 cells using a retroviral vector. We have constructed a series of Lu deletion mutants, expressed these in K562 cells, and used them to map monoclonal antibody binding sites and blood group antigens to individual Ig-like domains.8 9 We have defined the structure of the LU gene. This information has facilitated the elucidation of the molecular basis of the Lua/Lub and the Aua/Aub blood group polymorphisms.
MATERIALS AND METHODS
Antibodies and blood samples.Monoclonal antibodies against Lu gp BRIC 1087 (IgG1, anti-Lub), BRIC 221 (IgG2b), and BRIC 224 (IgG1)4 were as described. Monoclonal anti-Lub LM342/767.3110 was provided by Dr R. Fraser and Dr G. Inglis, Glasgow and West of Scotland Transfusion Service, Carluke, Scotland. Human sera containing alloantibodies to Lu system antigens were obtained, over many years, either through identification of unknown antibodies or as gifts from colleagues. Antibodies were prepared by adsorption onto antigen-positive red blood cells and subsequent elution at low pH (Elu-Kit II; Gamma Biologicals, Houston, TX). Blood samples from volunteer blood donors were obtained from the National Blood Service, Bristol and Oxford, UK.
Monoclonal antibody-specific immobilization of erythrocyte antigens (MAIEA) assay.MAIEA analysis, a competitive binding assay between mouse monoclonal antibodies and human alloantibodies, was performed as described previously.11 Trypsin-treated red blood cells12 were used as antigen-negative controls.
Molecular biology.All restriction enzymes and DNA modifying enzymes were obtained from commercial sources and used as recommended with the supplied buffers. DNA sequencing of plasmids and direct sequencing of polymerase chain reaction (PCR) products was performed using dye-labeled terminator chemistry on a 373A Applied Biosystems automated DNA sequencer (Perkin-Elmer, Warrington, UK). Genomic DNA was prepared from peripheral blood leukocytes as described.13LU genomic DNA corresponding to the complete open reading frame of Lu gp mRNA4 was amplified by PCR using the method of Cheng et al.14 Two overlapping PCR products were generated; the first, 3-kb product (nt 16 to 297 of Lu mRNA) was amplified using the sense primer Lu-gen9, CGTGAACATGGAGCCCCCGGACGCACC (nt 16-42), and the antisense primer Lu-gen5, ATTGTGACCTGGAGCTCAGAGCCCTGCAT (nt 297-269) and the second 8-kb product (nt 120-1936) using the sense primer Lu-gen1, TGCGCTTGTCTGTACCCCCGCTGGT (nt 120-144) and the antisense primer Lu-gen3, AGGGACAGCCTCTAGGAGGTTCTTGG (nt 1936-1911). DNA amplification products were purified by electrophoresis in 0.8% agarose gels and excised bands were extracted for sequence analysis using the Qiaex II gel extraction kit (Qiagen Ltd, Crawley, UK).
Cloning of full-length Lu cDNA and domain-deletion mutants into pBabe puro retroviral vector.Full-length Lu cDNA4 was assembled in pBluescript vector (Stratagene Ltd, Cambridge, UK) using sequences from clones containing the 5′ sequence (nts 1-180) and the remaining sequence (nts 26-2437) using a unique BssHII restriction site at nt 50. A major part of the 3′ noncoding sequence was removed using a unique Nae I site at nt 2048. This full-length Lu cDNA construct was subcloned into the pBabe puro retroviral vector (kindly provided by Dr H. Land, ICRF, London, UK). Domain-deletion mutants lacking the coding region(s) for one or more Ig-like domains were made using an inverse PCR strategy.15 Template was the full-length cDNA in pBluescript vector from which the EcoRI restriction site was removed by digestion with EcoRI, blunt-ending with S1 nuclease, and religation. Oligonucleotide PCR primers with in-frame, 5′ EcoRI sites were designed. A sense primer (Sdel5) 5′GGAATTCCCAGACCTCCCAGGCTG (nt 1647-1664), which straddles the coding region for the predicted C-terminal boundary of the fifth Ig-like domain of Lu gp, was used in conjunction with four antisense primers complimentary to the coding regions for the predicted N-terminal boundaries of the second, third, fourth, and fifth Ig-like domains: ASdel2 5′GGAATTCTCAGTGGCCTCTGGCTT (nt 475-458); ASdel3 5′GGAATTCACGTGCTCCGTGGGATA (nt 823-806); ASdel4 5′GGAATTCAGATAGGCCACGCGCAG (nt 1105-1088); and ASdel5 5′GGAATTCAGCTCTGGCGAGCCTTGG (nt 1371-1354). In each case, a pair of primers was used with Pfu polymerase (Stratagene Ltd, Cambridge, UK) to amplify a linear PCR product encoding the deleted Lu cDNA and the whole of the Bluescript vector sequence. These PCR products were digested with EcoRI and religated to form closed circular plasmids. The deleted Lu cDNA inserts were subcloned into pBabe puro retroviral vector and the DNA sequence of the cDNA inserts was verified. The constructs encoded the series of mutants depicted, together with the full-length Lu gp, in Fig 1. In each case, the deletion boundary was proximal to the predicted transmembrane region and included the amino acid residues NS encoded by the EcoRI restriction site.
Expression in K562 cells.The pBabe puro constructs were linearized by digestion with Sca I before transfection. K562 cells (5 × 105) were directly transfected with the pBabe puro construct containing full-length Lu cDNA (25 μg) using the calcium phosphate technique13 and with constructs containing domain-deleted Lu cDNA by electroporation (200V, 1,000 μF ). Transfectants were cultured in Iscove's modified Eagle's medium, 10% fetal calf serum (FCS) for 48 hours then plated over 96-well culture plates in medium containing 3 μg/mL puromycin (Sigma, Poole, UK). Individual puromycin-resistant colonies were isolated and tested for expression of Lu gp or domain-deletion mutants using murine monoclonal antibodies or eluates prepared from blood group antisera by flow cytometry as described.16 Nontransfected K562 cells, K562 cells transfected with “empty” pBabe puro vector, and K562 cells transduced with cDNA encoding the Fyb blood group gp16 were tested as negative controls. In tests with monoclonal antibodies, the mean fluorescence intensity (MFI) of the cells was recorded. In tests with human blood group alloantibodies, relative mean fluorescence (RMF = ratio of MFI of cells incubated with antibody/MFI of cells incubated without antibody) was calculated. K562 clones were also tested by immunoblotting as described.17 Membrane proteins were extracted from K562 cells using 1% (wt/vol) Triton X-100 (Sigma, Poole, UK) as described.18 Control red blood cell membranes were as described.17
RESULTS
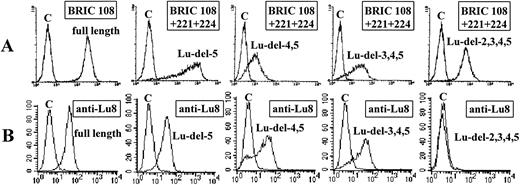
Analysis of K562 clones transfected with full-length and domain-deleted Lu cDNA.K562 clones transfected with full-length Lu cDNA as described in Materials and Methods were tested for surface expression of Lu gp by flow cytometric analysis using murine monoclonal antibodies against Lu gp (BRIC 108, BRIC 221, and BRIC 224). All three antibodies bound strongly to positive clones. The result obtained using the monoclonal anti-Lub BRIC 108 is shown in Fig 2A (extreme left-hand histogram). BRIC 221 and BRIC 224 gave very similar results (data not shown). Nontransfected K562 cells, K562 cells transfected with “empty” pBabe puro vector, and K562 cells transduced with cDNA encoding the Fyb blood group gp tested with monoclonal antibodies BRIC 108, BRIC 221, and BRIC 224 all gave similar MFI values and were negative.
Flow cytometric analysis of K562 transfectants expressing full-length and domain-deleted Lu cDNA. (A) Transfectants tested with monoclonal antibodies against Lu gp. (B) Transfectants tested with human anti-Lu8. The number of cells is plotted on the ordinate and the fluorescence intensity is plotted on the abscissa. C denotes control cells: K562 cells transfected with Fyb glycoprotein in the test with BRIC 108, untransfected K562 cells in all other tests.
Flow cytometric analysis of K562 transfectants expressing full-length and domain-deleted Lu cDNA. (A) Transfectants tested with monoclonal antibodies against Lu gp. (B) Transfectants tested with human anti-Lu8. The number of cells is plotted on the ordinate and the fluorescence intensity is plotted on the abscissa. C denotes control cells: K562 cells transfected with Fyb glycoprotein in the test with BRIC 108, untransfected K562 cells in all other tests.
K562 cells transfected with the domain-deleted Lu cDNAs Lu-del-5; Lu-del-4,5; Lu-del-3,4,5 and Lu-del-2,3,4,5 (Fig 1) were examined for expression by flow cytometry using a pool of BRIC 108, BRIC 221, and BRIC 224. Clones were obtained that expressed each of the four domain-deletion mutants and each of these gave strong fluorescence signals (Fig 2A). Each clone was then retested using the monoclonal antibodies individually. The results are summarized in Table 1. BRIC 108 (anti-Lub) and BRIC 224 bound to all of the domain-deletion mutants in addition to the clone expressing the full-length Lu gp. The epitopes recognized by these antibodies are, therefore, expressed on the first, N-terminal domain of the Lu gp. In contrast, BRIC 221 bound to the clone expressing full-length Lu gp and to the Lu-del-5 expressing clone, but failed to bind to clones expressing deleted Lu gp, which lacked the fourth domain (Lu-del-4,5; Lu-del-3,4,5 and Lu-del-2,3,4,5). The BRIC 221 epitope is, therefore, carried on the fourth domain of Lu gp.
Flow Cytometric Analysis of K562 Transfectants Expressing Full-Length and Domain-Deleted Lu cDNA
| K562 Cells Transfected With cDNA Constructs . | Monoclonal Antibodies . | Human Alloantibodies . | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | BRIC 108 . | BRIC 224 . | BRIC 221 . | Lub . | Lu5 . | Lu12 . | Lu4 . | Lu8 . | Lu20 . | Lu6 . | Lu7 . | Aub . | Lu13 . |
| Full-length Lu cDNA | 19 | 96 | 114 | 17 | 8 | 6 | 21 | 10 | 12 | 6 | 10 | 11 | 5 |
| Lu-del-5 | 33 | 143 | 196 | 14 | 5 | 4 | 15 | 7 | 7 | 6 | 5 | 1 | 1 |
| Lu-del-4,5 | 17 | 59 | 5.0 | 21 | 8 | 6 | 20 | 11 | 11 | 2 | 1 | 2 | 1 |
| Lu-del-3,4,5 | 17 | 62 | 2.6 | 18 | 8 | 6 | 21 | 9 | 1 | 1 | 2 | 1 | 1 |
| Lu-del-2,3,4,5 | 15 | 227 | 7.4 | 13 | 5 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Untransfected K562 cells (control) | 3.6 | 3.0 | 3.0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 |
| Predicted domain location of antigen/epitope | 1 | 1 | 4 | 1 | 1 | 1 or 2 | 2 | 2 | 3 | 3 or 4 | 4 | 5 | 5 |
| K562 Cells Transfected With cDNA Constructs . | Monoclonal Antibodies . | Human Alloantibodies . | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | BRIC 108 . | BRIC 224 . | BRIC 221 . | Lub . | Lu5 . | Lu12 . | Lu4 . | Lu8 . | Lu20 . | Lu6 . | Lu7 . | Aub . | Lu13 . |
| Full-length Lu cDNA | 19 | 96 | 114 | 17 | 8 | 6 | 21 | 10 | 12 | 6 | 10 | 11 | 5 |
| Lu-del-5 | 33 | 143 | 196 | 14 | 5 | 4 | 15 | 7 | 7 | 6 | 5 | 1 | 1 |
| Lu-del-4,5 | 17 | 59 | 5.0 | 21 | 8 | 6 | 20 | 11 | 11 | 2 | 1 | 2 | 1 |
| Lu-del-3,4,5 | 17 | 62 | 2.6 | 18 | 8 | 6 | 21 | 9 | 1 | 1 | 2 | 1 | 1 |
| Lu-del-2,3,4,5 | 15 | 227 | 7.4 | 13 | 5 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Untransfected K562 cells (control) | 3.6 | 3.0 | 3.0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 |
| Predicted domain location of antigen/epitope | 1 | 1 | 4 | 1 | 1 | 1 or 2 | 2 | 2 | 3 | 3 or 4 | 4 | 5 | 5 |
In tests with monoclonal antibodies mean fluorescence intensity (MFI) is given. In tests with human blood group alloantibodies relative mean fluorescence (RMF) calculated as described in Materials and Methods is given.
The same transfectants were tested by flow cytometry with human antibodies to the Lu blood group antigens Lub (Lu2); Lu4; Lu5; Lu6; Lu7; Lu8; Lu12; Lu13; Aua (Lu18), Aub (Lu19) and Lu20. The results obtained with anti-Lu8 are shown in Fig 2B and the results obtained with other antibodies are summarized in Table 1. Anti-Lub and anti-Lu5 reacted with all transfectants suggesting that the antigens they recognize are on the first (N-terminal) domain. Anti-Lu4 and anti-Lu8 reacted with all transfectants except those lacking the second domain suggesting that the antigens they recognize are on domain 2, anti-Lu20 reacted with all except those lacking the third domain suggesting that Lu20 is on domain 3, and anti-Lu7 reacted with all except those lacking the fourth domain suggesting that Lu7 is on domain 4. Anti-Lu13 and anti-Aub were only reactive with the transfectant expressing the complete cDNA, suggesting that the antigens they recognize are on the fifth domain. Anti-Aua did not react with the transfectant expressing the complete glycoprotein as would be expected because Aua is allelic to Aub. Nontransfected K562 cells, K562 cells transfected with “empty” pBabe puro vector, and K562 cells transduced with cDNA encoding the Fyb blood group gp tested with the human antibodies were negative.
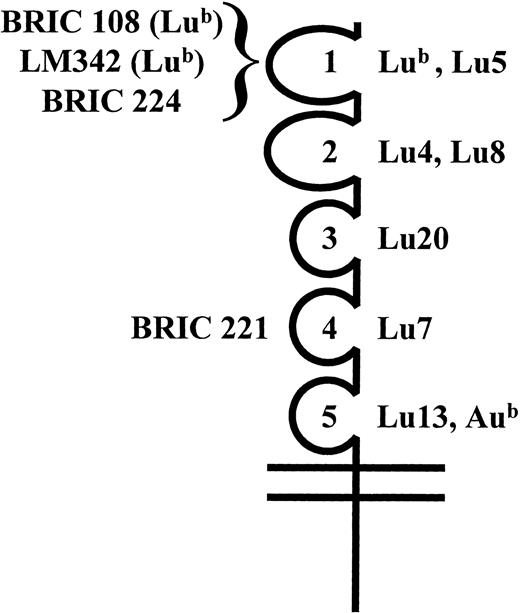
Results obtained with anti-Lu12 and anti-Lu6 were less clear cut (Table 1): anti-Lu6 reacted with those transfectants expressing the third domain, but the reaction with the cells lacking only domains 4 and 5 was weak; anti-Lu12 reacted with all transfectants, but the reaction with the cells only expressing the first domain was weak. Further evidence that the Lu12 antigen is expressed on domain 1 was obtained using a competitive binding assay (MAIEA analysis, Table 2). Human anti-Lu12 and anti-Lu5 blocked the binding of two mouse monoclonal anti-Lub antibodies (BRIC 108 and LM342) although neither anti-Lu12 nor anti-Lu5 blocked the binding of BRIC 224, which also recognizes an epitope on domain 1. By contrast, human anti-Lub blocked the binding of BRIC 108, LM342, and BRIC 224. A summary of the suggested locations of the Lu system antigens and the epitopes recognized by monoclonal antibodies is shown in Fig 3.
Results of a MAIEA Analysis Shown as a Ratio of Absorbance Obtained With Antigen Positive Red Blood Cells to That Obtained With Antigen Negative Cells
| Mouse Monoclonal Antibodies . | Domain . | Human Alloantibodies . | |||||||
|---|---|---|---|---|---|---|---|---|---|
| . | . | Lub . | Lu5 . | Lu6 . | Lu8 . | Lu12 . | Lu13 . | Aub . | Lu20 . |
| Anti-Lub BRIC 108 | 1 | 1.0 | 1.3 | 9.5 | 22.0 | 1.1 | 5.3 | 9.7 | 8.5 |
| Anti-Lub LM342 | 1 | 1.1 | 1.9 | 11.4 | 19.3 | 1.3 | 6.2 | 24.6 | 10.0 |
| BRIC 224 | 1 | 1.3 | 3.7 | 4.2 | 18.8 | 13.0 | 4.5 | 18.7 | 5.5 |
| BRIC 221 | 4 | 4.1 | 4.2 | 3.2 | 8.9 | 10.8 | 1.9 | 4.0 | 3.0 |
| Mouse Monoclonal Antibodies . | Domain . | Human Alloantibodies . | |||||||
|---|---|---|---|---|---|---|---|---|---|
| . | . | Lub . | Lu5 . | Lu6 . | Lu8 . | Lu12 . | Lu13 . | Aub . | Lu20 . |
| Anti-Lub BRIC 108 | 1 | 1.0 | 1.3 | 9.5 | 22.0 | 1.1 | 5.3 | 9.7 | 8.5 |
| Anti-Lub LM342 | 1 | 1.1 | 1.9 | 11.4 | 19.3 | 1.3 | 6.2 | 24.6 | 10.0 |
| BRIC 224 | 1 | 1.3 | 3.7 | 4.2 | 18.8 | 13.0 | 4.5 | 18.7 | 5.5 |
| BRIC 221 | 4 | 4.1 | 4.2 | 3.2 | 8.9 | 10.8 | 1.9 | 4.0 | 3.0 |
The location of monoclonal antibody epitopes and blood group antigens on the five IgSF domains of Lu gp. Monoclonal antibody epitopes BRIC 108, BRIC 221, BRIC 224, and LM342 and blood group antigens Lub, Lu4, Lu5, Lu7, Lu8, Lu13, Aub, and Lu20 were mapped to individual IgSF domains as indicated.
The location of monoclonal antibody epitopes and blood group antigens on the five IgSF domains of Lu gp. Monoclonal antibody epitopes BRIC 108, BRIC 221, BRIC 224, and LM342 and blood group antigens Lub, Lu4, Lu5, Lu7, Lu8, Lu13, Aub, and Lu20 were mapped to individual IgSF domains as indicated.
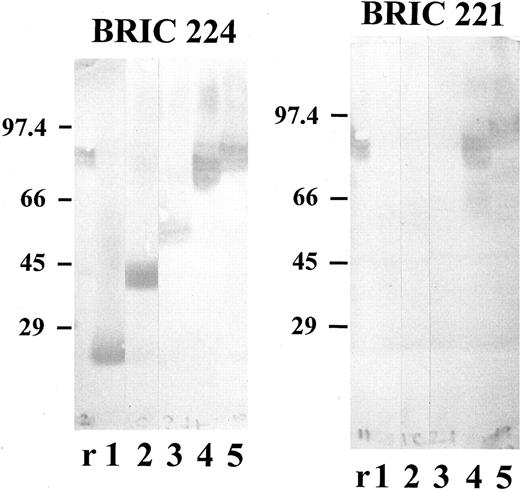
Triton extracts (prepared as described in Materials and Methods) from the transfected K562 cells were tested by immunoblotting with each of the monoclonal antibodies. The results (Fig 4) were consistent with those obtained by flow cytometric analysis. BRIC 108 (data not shown) and BRIC 224 (Fig 4) bound to all of the domain-deletion mutants, as well as to full-length Lu gp, whereas BRIC 221 bound only to full-length Lu gp and to the Lu-del-5 mutant (Fig 4). The Lu gp expressed by K562 cells transfected with full-length Lu cDNA migrated with Mr 86,000. The apparent molecular weights of the domain-deleted proteins were: Lu-del-5 - Mr 78,500; Lu-del-4,5 - Mr 57,500; Lu-del-3,4,5 - Mr 43,500; Lu-del-2,3,4,5 - Mr 24,000.
Immunoblotting of K562 transfectants expressing full-length and domain-deleted Lu cDNA. Triton extracts prepared from K562 transfectants as described in Materials and Methods and solubilized red blood cell membranes were separated on 10% (wt/vol) polyacrylamide gels and probed with monoclonal antibodies BRIC 224 or BRIC 221 by Western blotting. r, red cell membranes; 1, Lu-del-2,3,4,5 transfectant; 2, Lu-del-3,4,5 transfectant; 3, Lu-del-4,5 transfectant; 4, Lu-del-5 transfectant; 5, Lu-full-length transfectant. Mr values for the stained bands were calculated from the migration of the indicated standard proteins.
Immunoblotting of K562 transfectants expressing full-length and domain-deleted Lu cDNA. Triton extracts prepared from K562 transfectants as described in Materials and Methods and solubilized red blood cell membranes were separated on 10% (wt/vol) polyacrylamide gels and probed with monoclonal antibodies BRIC 224 or BRIC 221 by Western blotting. r, red cell membranes; 1, Lu-del-2,3,4,5 transfectant; 2, Lu-del-3,4,5 transfectant; 3, Lu-del-4,5 transfectant; 4, Lu-del-5 transfectant; 5, Lu-full-length transfectant. Mr values for the stained bands were calculated from the migration of the indicated standard proteins.
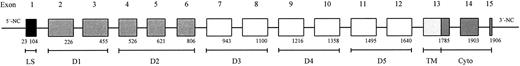
Structure of the LU gene and the molecular basis of the Lua/Lub and Aua/Aub polymorphisms.To determine the molecular basis of Lu blood group polymorphisms, the structure of the LU gene corresponding to the open reading frame of Lu mRNA was defined and the nucleotide sequences of the intron/exon boundaries were determined (Table 3). This region of the LU gene comprises 15 exons spanning approximately 11 kb (Fig 5). Domains 1, 3, 4, and 5 are each encoded by two exons and domain 2 by three exons.
Sequence of LU Intron/Exon Boundaries, Exon Size, and Approximate Intron Size
| Exon . | Domain . | 3′ End of Intron Exon 5′ End of Intron . | Size of Exon (bp) . | Size of Intron (kb) . |
|---|---|---|---|---|
| 1 | Lu5′UT/LS | ... GCGCACCCAG gtatggctcg | >823-150 | 2.0 |
| 2 | D1a | tctcttgcag ATGCCCAGGC ... ... ATGGTTCCTT gtgagtgctt | 122 | 0.7 |
| 3 | D1b | gcgcccacag ACCGACCGCT ... ... AACGTGTTTG gtaagtgtcc | 229 | 0.09 |
| 4 | D2a | tttttcacag CAAAGCCAGA ... ... TGCCCAGGAG gtacctctcg | 71 | 0.5 |
| 5 | D2b | tgcccaggag ATCGCCACCT ... ... ATGAACCCAG gtgagcagcg | 95 | 0.09 |
| 6 | D2c | tctctcccag AGGGCTACAT ... ... ACCCTGCACT gtgagtctgt | 185 | 0.53 |
| 7 | D3a | cttcccttag ATCCCACGGA ... ... CCGCCTTCAG gtgacccacc | 137 | 0.31 |
| 8 | D3b | cgtctcccag GATGAGCAGG ... ... CGCGTGGCCT gtgagagccc | 157 | 3.5 |
| 9 | D4a | cccactgcag ATCTGGACCC ... ... CTGGACCAAG gtgacagga | 116 | 0.1 |
| 10 | D4b | cccttcccag GACTCCACTC ... ... CTGGTCCAAG gttcaggggg | 142 | 0.17 |
| 11 | D5a | ccacccacag GCTCGCCAGA ... ... GGGGGGCAGC gtaccacttc | 137 | 0.15 |
| 12 | D5b | gcctccatag CCCGCAGAGC ... ... TTCGGCGCCG gtgagtgact | 145 | 0.09 |
| 13 | TM | gtccctgcag TGAGCCCCCA ... ... AGGGGGCTCC gtgagtggcc | 145 | 0.97 |
| 14 | TM/Cyto | ctccccccag GCCGCCAGGG .. ... CGGAGACGAG gtgggtgagg | 118 | 0.09 |
| 15 | Cyto/3′UT | cgctccccag TGCTGATGAG .. | >33-150 |
| Exon . | Domain . | 3′ End of Intron Exon 5′ End of Intron . | Size of Exon (bp) . | Size of Intron (kb) . |
|---|---|---|---|---|
| 1 | Lu5′UT/LS | ... GCGCACCCAG gtatggctcg | >823-150 | 2.0 |
| 2 | D1a | tctcttgcag ATGCCCAGGC ... ... ATGGTTCCTT gtgagtgctt | 122 | 0.7 |
| 3 | D1b | gcgcccacag ACCGACCGCT ... ... AACGTGTTTG gtaagtgtcc | 229 | 0.09 |
| 4 | D2a | tttttcacag CAAAGCCAGA ... ... TGCCCAGGAG gtacctctcg | 71 | 0.5 |
| 5 | D2b | tgcccaggag ATCGCCACCT ... ... ATGAACCCAG gtgagcagcg | 95 | 0.09 |
| 6 | D2c | tctctcccag AGGGCTACAT ... ... ACCCTGCACT gtgagtctgt | 185 | 0.53 |
| 7 | D3a | cttcccttag ATCCCACGGA ... ... CCGCCTTCAG gtgacccacc | 137 | 0.31 |
| 8 | D3b | cgtctcccag GATGAGCAGG ... ... CGCGTGGCCT gtgagagccc | 157 | 3.5 |
| 9 | D4a | cccactgcag ATCTGGACCC ... ... CTGGACCAAG gtgacagga | 116 | 0.1 |
| 10 | D4b | cccttcccag GACTCCACTC ... ... CTGGTCCAAG gttcaggggg | 142 | 0.17 |
| 11 | D5a | ccacccacag GCTCGCCAGA ... ... GGGGGGCAGC gtaccacttc | 137 | 0.15 |
| 12 | D5b | gcctccatag CCCGCAGAGC ... ... TTCGGCGCCG gtgagtgact | 145 | 0.09 |
| 13 | TM | gtccctgcag TGAGCCCCCA ... ... AGGGGGCTCC gtgagtggcc | 145 | 0.97 |
| 14 | TM/Cyto | ctccccccag GCCGCCAGGG .. ... CGGAGACGAG gtgggtgagg | 118 | 0.09 |
| 15 | Cyto/3′UT | cgctccccag TGCTGATGAG .. | >33-150 |
Abbreviations: Lu5′UT, Lu 5′untranslated sequence; LS, leader sequence; D, IgSF domain; TM, membrane-spanning domain; Cyto, cytoplasmic domain; Lu3′UT, Lu 3′untranslated sequence.
Size of exon shows number of bases in coding sequence only.
Schematic representation of the LU gene showing exons and the corresponding structural domains of Lu gp. D1-5 are IgSF domains. In the mRNA species encoding the Lu gp isoform B-CAM, the intron between exons 13 and 14 is not spliced.8 The numbering of nucleotides is as given in the Genbank database entry for Lu gp cDNA, accession number X83425, abbreviations are as in Table 3.
Schematic representation of the LU gene showing exons and the corresponding structural domains of Lu gp. D1-5 are IgSF domains. In the mRNA species encoding the Lu gp isoform B-CAM, the intron between exons 13 and 14 is not spliced.8 The numbering of nucleotides is as given in the Genbank database entry for Lu gp cDNA, accession number X83425, abbreviations are as in Table 3.
Flow cytometric analysis of K562 transfectants, described above, suggested that the Lub antigen was defined by the amino acid sequence of the first domain of Lu gp. Genomic DNA from 4 unrelated individuals of the Lu(a+b−) phenotype, 2 unrelated individuals of the Lu(a−b+) phenotype, and 2 unrelated individuals of the Lu(a+b+) phenotype was amplified by PCR and the sequence of exons 2 and 3 (corresponding to the cDNA sequence of domain 1, Fig 5) determined. A single base change in exon 3 (G252 to A) was found in genomic DNA from all six Lu(a+) individuals, with mixed sequences present in the two heterozygotes. This results in a predicted Arg77 (Lub ) to His (Lua) amino acid change in Lu gp.
Flow cytometric analysis of K562 transfectants suggested that the Aub antigen was located on domain 5, which is encoded by exons 11 and 12 (Fig 5). Sequence analysis of these exons in genomic DNA amplified from 2 unrelated individuals of the Au(a−b+) phenotype, 3 unrelated individuals with the Au(a+b−) phenotype, and 2 unrelated individuals with the Au(a+b+) phenotype showed a single base substitution (G1637 to A) in exon 12 resulting in the predicted change Ala539 (Aub) to Thr (Aua) in Lu gp.
DISCUSSION
Expression in K562 cells confirms the identity of the Lu cDNA (which was isolated from a placental cDNA library) as cDNA for the Lutheran blood group active glycoprotein. Testing of domain-deletion mutants has mapped many of the Lu blood group antigens to individual, extracellular, IgSF domains of this molecule (Fig 3). The results obtained with anti-Lu6 and with anti-Lu12 were, however, not conclusive. It is possible that the removal of certain domains may affect the conformation of adjacent domains and thus the expression of some antigens. It appears from the results that the Lu6 antigen may be on domain 3, but expressed only weakly in the absence of the fourth domain and that Lu12 may be on domain 1, but not fully expressed in the absence of domain 2. It is also possible that the accessibility of the Lu12 antigen for antibody binding on the domain-deletion mutant containing only domain 1 is affected by the position of domain 1 close to the membrane in this mutant (Fig 1) and that the Lu6 antigen in the domain-deletion mutant lacking domains 4 and 5 might be similarly affected. Although outside the scope of the present study, it would be possible to address such questions by testing cells expressing chimeric constructs containing individual domains of Lu gp together with one or more irrelevant IgSF domains, inserted as “spacers.” The use of such chimeric constructs will be of particular importance in functional studies. The results obtained in MAIEA analysis are unaffected by these constraints and, as discussed below, provided independent evidence for the location of Lu12 on domain 1.
It is interesting to note that the monoclonal antibodies BRIC 108 and BRIC 224 and human anti-Lua and anti-Lub, which define epitopes/antigens on the N-terminal domain, directly agglutinate red blood cells in saline medium. By analogy with the predicted size of the C2-set domains of CD58,19 the Lu gp structure would be predicted to extend ≈17.5 nm from the membrane lipid bilayer. Thus, the N-terminal domain of Lu gp would be presented outside the red blood cell membrane glycocalyx, which is thought to extend ≈12.5 nm from the bilayer.20,21 BRIC 221, which recognizes an epitope on the fourth domain, agglutinates red blood cells only in an indirect antiglobulin technique, and this may be related to the inability of IgG antibodies to span the distance between red blood cells in suspension when the epitope is close to the lipid bilayer rather than above the glycocalyx.22
The exons of the LU gene correlate with the predicted domain structure of the Lu gp with domains 1, 3, 4, and 5 encoded by two exons and domain 2 by three exons. All intron-exon boundaries conform to the GT/AT sequence rule.23 The Lua and Lub antigens are defined by an Arg77 (Lub ) to His (Lua) amino acid substitution in the predicted C′ strand on the CFG face of the first, V-set, Ig-like domain. The Aua/Aub polymorphism was found to be determined by an amino acid substitution Ala539 (Aub) to Thr (Aua) in the G strand of the fifth, C2-set, Ig-like domain. Anti-Aua and anti-Aub are rare antibodies, usually only found in sera containing several antibodies to red blood cell antigens,1 despite about 20% and 50% of Europeans lacking Aua and Aub, respectively. This suggests that the Aua and Aub antigens are less immunogenic than many other blood group antigens, a feature that may be related to the nature of the amino acids that define these antigens. The results of MAIEA assays demonstrated competition between human anti-Lub and the mouse antibody BRIC 224, consistent with the location of the Lub antigen and the BRIC 224 epitope on the N-terminal domain. There was also competition between mouse monoclonal anti-Lub antibodies and human anti-Lu5 and anti-Lu12, supporting the mapping of these antigens to domain 1. In contrast, there was no competition in tests using BRIC 224 (which also maps to domain 1) with human anti-Lu5 or anti-Lu12. Taken together, these results suggest that the Lub antigen is located near the BRIC 224 epitope and Lu5 and Lu12 antigens, but that the BRIC 224 epitope is sufficiently remote from the Lu5 and Lu12 antigens such that blocking does not occur. Similar results were obtained from MAIEA analyses using monoclonal antibodies against CD55 and human alloantibodies in the Cromer blood group system. The Tca and WESb antigens of the Cromer system are both defined by amino acid residues on the N-terminal short consensus repeat of CD55, a glycoprotein of the complement control superfamily,24,25 yet monoclonal antibodies that block anti-Tca in MAIEA analysis do not block anti-WESb and monoclonal antibodies that block anti-WESb do not block anti-Tca.26 27
The Lu gp domain-deletion mutants were studied by immunoblotting and values for relative molecular mass were calculated. Comparison of the Mr calculated for each C2 set domain was consistent with the predicted glycosylation pattern of Lu gp, ie, deletion of the fifth (C2-set) domain, which lacks N-glycosylation sequence motifs resulted in a change in Mr of 7,500; deletion of the fourth (C2-set) domain, which has four potential N-glycosylation sites, resulted in a change in Mr of 21,000 and deletion of the third (C2 set) Domain, which has a single potential N-glycosylation site, resulted in a change in Mr of 14,000. These results clearly indicate that the N-glycosylation site on the third domain is used when the protein is expressed in K562 cells and that one or more of the four potential N-glycosylation sites on the fourth domain is/are also used under these conditions.
Other IgSF molecules found on mature red blood cells are the LW gp, an intercellular adhesion molecule (ICAM) homologue28,29 that is reported to bind leukocyte function antigen-1 (LFA-1),30 CD58,31 CD47,32,33 and the Oka blood group gp.34 LW gp has one known blood group mutation resulting in an amino acid substitution on the ABE face of the first domain,28,35 the opposite face to that defined as the integrin binding face in other members of the ICAM family. This mutation occurs in up to 8% of Estonians, but is rare outside Northern Europe. Red blood cell membrane CD58 and CD47 have no known blood group mutations.31 The Oka blood group gp has one known blood group mutation, which is very rare and so far found only in Japanese.34 In contrast, the Lu gp clearly has several blood group mutations. We have shown in this study that at least one Lutheran antigen is located on each of the five IgSF domains, a unique feature among the known IgSF molecules. The adhesive and/or receptor properties of the Lu gp on hematopoietic and other tissues are, as yet, undefined. The location of blood group antigens on each domain of the Lu gp and thereby the availability of antibodies to sites on each of the five IgSF domains should prove to be useful in studies of the function of this molecule, should an adhesive or receptor ligand be identified.
NOTE ADDED IN PROOF
Following submission of this manuscript Udani et al36 presented evidence that the Lu gp binds the extracellular matrix protein laminin.
ACKNOWLEDGMENT
We thank Peter Martin, IBGRL, Bristol, UK, for performing DNA sequence analyses.
Presented in part at the 49th Annual Congress of the American Association of Blood Banks, Orlando, FL, October 12-16, 1996.
Address reprint requests to S.F. Parsons, PhD, Bristol Institute for Transfusion Sciences, Southmead Rd, Bristol, BS10 5ND, UK.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal