Abstract
Chronic lymphocytic leukemia (CLL) is a B-cell tumor involving small lymphocytes that generally express the CD5 antigen and low levels of surface Ig. Within this definition, there is heterogeneity among cases in cell morphology, karyotypic abnormalities, and clinical course. Trisomy 12, the most frequent karyotypic abnormality, is commonly found in a subset of CLL with atypical morphology. It has also been associated with advanced disease, and possibly with a less favorable prognosis. A further subset of cases with abnormalities involving chromosome 13q14 have typical lymphocyte morphology. Occasionally, the two abnormalities are found together. To assess the clonal history of the cell of origin in disease subsets defined by these two chromosomal abnormalities, we investigated the usage of VH genes and the pattern of somatic mutation in 10 cases of trisomy 12 with atypical morphology and eight cases of 13q14 abnormality with typical morphology. In addition, four cases with both chromosomal abnormalities were analyzed. Results confirm a common usage of the VH1 family in all subsets. However, the patterns of somatic mutation were distinct, with cases of trisomy 12 showing a minimal level of mutation (mean ± SD, 0.34% ± 0.86%) and cases of 13q14 abnormality showing significant levels (6.5% ± 1.67%). The four cases with both abnormalities showed a mixed pattern. All mutated cases had intraclonal homogeneity, and three of 10 had a pattern indicative of antigen selection. These results suggest that the clonal history of the two subsets of CLL may differ.
CHRONIC LYMPHOCYTIC leukemia (CLL) is a B-cell tumor with a characteristic phenotype. However, heterogeneity within CLL is exhibited both in the presenting clinical features and in the rate of disease progression.1 An adverse prognosis appears to correlate with chromosomal abnormalities and atypical morphology of the tumor cells. The most frequent chromosome abnormalities in CLL are trisomy 12, found in 15% to 18% of cases, and deletions or translocations of chromosome 13q14, found in 17% to 29%.2 The latter group has the same median survival as those with a normal karyotype, whereas trisomy 12 has been associated with more advanced disease and may act as a marker of poorer prognosis.3
Atypical morphology is a term applied to cases in which cells with features of prolymphocytes, or a pleomorphic cell population (including large lymphocytes, cells with clefted nuclei, or cells with lymphoplasmacytoid features), comprise greater than 10% of blood lymphocytes.4 The association between an increased number of prolymphocytes and a poor prognosis is well documented. More recently, it has been found that atypical lymphocyte morphology is frequently accompanied by trisomy 12.5-8 It has also been confirmed in a large study of 544 patients that trisomy 12 defines a subgroup of CLL with a more advanced disease stage and possibly worse prognosis.8 The role of chromosomal changes in the pathogenesis of CLL is unclear. Trisomy 12 or 13q14 abnormalities as detected by chromosome analysis, fluorescence in situ hybridization (FISH), or molecular methods are commonly found as the sole genetic abnormality in early-stage disease. However, trisomy 12 is found in only a proportion of the tumor population, indicating that it is unlikely to be an initiating factor in tumorigenesis.6 Similarly, deletion of the retinoblastoma gene assigned to chromosome 13q14 is only found in a percentage of the malignant clone.9 Although trisomy 12 or 13q14 abnormalities are found separately in a substantial proportion of cases of CLL, they coexist in only 2% to 5% of patients, implying that each change may be involved in a distinct pathogenic route.
The ability to amplify and sequence the Ig heavy and light chain variable region genes has provided a new and powerful method for investigating B-cell malignancies. It is now clear that there are approximately 51 VH genes in the repertoire available for recombination to generate a functional VH-DH-JH transcriptional unit.10 Studies of gene usage in CLL have indicated that there may be asymmetry in the choice of VH and VL genes used in the cell of origin (reviewed in Kipps11 ). Nonrandom differential usage of VH genes has also been observed for other B-cell tumors.12 13
Following genetic recombination, a normal B cell may undergo stimulation by antigen and enter the lymphoid follicle. At this point in differentiation, the somatic hypermutation mechanism is activated, introducing point mutations into the variable region genes (reviewed in Berek and Milstein14 and Gray15 ). In the presence of limiting antigen, there will be subsequent selection of V gene sequences that provide increased affinity for antigen, and this often leads to a concentration of replacement amino acids in the complementarity-determining regions (CDRs) (reviewed in Kocks and Rajewsky16 ).
The availability of a complete map of the VH gene repertoire and the enlarging database of expressed VH gene sequences means that it is now possible to assess the degree of somatic mutation in an expressed sequence with confidence.10 For CLL, there have been several small studies indicating that the level of somatic mutation in CLL is low (reviewed in Kipps11 and Schroeder and Dighiero17 ), and this has led to the conclusion that the cell of origin has not encountered antigen. However, there is a clear subset of cases in which there appears to be a significant degree of deviation from the germ line sequence indicative of somatic mutation.17 In view of this apparent heterogeneity, we decided to investigate cases of CLL that were defined clinically and by typical phenotypic markers. To link the investigation with described subsets of disease, we chose cases that were known to carry the major chromosomal abnormalities characteristic of CLL, trisomy 12 or 13q14.
Clinical Features of the Patients
| Patient No. . | Sex . | Age (yr) . | WBC Count (x 10−9/L) . | Morphology . | Stage . | Disease . | Therapy . |
|---|---|---|---|---|---|---|---|
| 1 | F | 51 | 10.5 | A | A | P | + |
| 2 | M | 58 | 13.4 | A | B | — | + |
| 3 | F | 67 | 47.2 | A | B | — | + |
| 4 | M | 55 | 12.8 | A | B | — | + |
| 5 | M | 71 | 25.8 | A | A | S | — |
| 6 | M | 76 | 16.3 | A | A | P | + |
| 7 | F | 81 | 14.9 | A | A | S | — |
| 8 | M | 84 | 25.4 | A | A | P | + |
| 9 | M | 79 | 12.7 | A | A | S | — |
| 10 | M | 90 | 12.8 | A | B | — | + |
| 11 | M | 46 | 28.9 | T | A | S | — |
| 12 | F | 78 | 15.8 | T | A | S | — |
| 13 | M | 56 | 8.1 | T | A | S | — |
| 14 | M | 73 | 19.1 | T | A | P | + |
| 15 | F | 73 | 13.3 | T | A | S | — |
| 16 | F | 78 | 20.1 | T | A | S | — |
| 17 | F | 43 | 16.6 | T | A | P | + |
| 18 | M | 62 | 43.0 | T | B | — | + |
| 19 | M | 73 | 16.6 | A | A | P | + |
| 20 | F | 67 | 25.8 | A | A | P | + |
| 21 | F | 78 | 12.2 | A | A | S | — |
| 22 | F | 60 | 12.1 | A | A | S | — |
| Patient No. . | Sex . | Age (yr) . | WBC Count (x 10−9/L) . | Morphology . | Stage . | Disease . | Therapy . |
|---|---|---|---|---|---|---|---|
| 1 | F | 51 | 10.5 | A | A | P | + |
| 2 | M | 58 | 13.4 | A | B | — | + |
| 3 | F | 67 | 47.2 | A | B | — | + |
| 4 | M | 55 | 12.8 | A | B | — | + |
| 5 | M | 71 | 25.8 | A | A | S | — |
| 6 | M | 76 | 16.3 | A | A | P | + |
| 7 | F | 81 | 14.9 | A | A | S | — |
| 8 | M | 84 | 25.4 | A | A | P | + |
| 9 | M | 79 | 12.7 | A | A | S | — |
| 10 | M | 90 | 12.8 | A | B | — | + |
| 11 | M | 46 | 28.9 | T | A | S | — |
| 12 | F | 78 | 15.8 | T | A | S | — |
| 13 | M | 56 | 8.1 | T | A | S | — |
| 14 | M | 73 | 19.1 | T | A | P | + |
| 15 | F | 73 | 13.3 | T | A | S | — |
| 16 | F | 78 | 20.1 | T | A | S | — |
| 17 | F | 43 | 16.6 | T | A | P | + |
| 18 | M | 62 | 43.0 | T | B | — | + |
| 19 | M | 73 | 16.6 | A | A | P | + |
| 20 | F | 67 | 25.8 | A | A | P | + |
| 21 | F | 78 | 12.2 | A | A | S | — |
| 22 | F | 60 | 12.1 | A | A | S | — |
Abbreviations: A, atypical; T, typical; P, progressive; S, stable; WBC, white blood cell.
Phenotypic and Karyotypic Analysis of Tumor Cells
| Patient No. . | Surface Markers . | Partial Karyotype . | ||||
|---|---|---|---|---|---|---|
| . | Ig . | CD5 . | CD23 . | CD22 . | FMC7 . | . |
| 1 | MDλ+ | + | + | + | − | +12 |
| 2 | MDκ+ | + | + | + | + | +12, del 14 (q11; q2?) |
| 3 | MDκ+ | + | + | + | + | +12 |
| 4 | MDκ+ | + | + | + | − | +12 |
| 5 | Mλ+ wk | + | + | + | + | +12, +18, +19 |
| 6 | MDκ+ | + | + | + | − | +12 |
| 7 | MDκ+ | + | + | + | + | +12 |
| 8 | MDκ+ | + | + | + | + | +12 |
| 9 | MDκ+ | + | + | − | − | +12 |
| 10 | MDκ+ | + | + | + | − | +12 |
| 11 | MDκ+ | + | + | + | − | del 13(q12; q14) |
| 12 | MD+ wk | + | + | − | − | del 13(q12; q14) |
| 13 | MDλ+ | + | + | − | − | del 13(q12; q22) |
| 14 | MDκ+ wk | + | + | − | − | t(13; 18)(q14; q22) |
| 15 | Mλ+ | + | + | + | − | t(13; 17)(q14; q11.2) |
| 16 | MDκ+ wk | + | + | − | − | t(13; 13)(q14; q22) |
| 17 | MDλ+ wk | + | + | + | − | del 13 (q14; q22) |
| 18 | Mλ+ wk | + | + | − | − | t(9; 13)(p11; q14) |
| 19 | Dκ+ wk | + | + | + | − | +12, del 13 (q12; q14) |
| 20 | MDκ+ | + | + | − | − | +12, t(12; 13)(q13; q14) |
| 21 | M+ wk | + | + | − | − | +12, del 13(q12; 14) |
| 22 | Mλ+ wk | + | + | + | − | +12, t(1; 13) |
| Patient No. . | Surface Markers . | Partial Karyotype . | ||||
|---|---|---|---|---|---|---|
| . | Ig . | CD5 . | CD23 . | CD22 . | FMC7 . | . |
| 1 | MDλ+ | + | + | + | − | +12 |
| 2 | MDκ+ | + | + | + | + | +12, del 14 (q11; q2?) |
| 3 | MDκ+ | + | + | + | + | +12 |
| 4 | MDκ+ | + | + | + | − | +12 |
| 5 | Mλ+ wk | + | + | + | + | +12, +18, +19 |
| 6 | MDκ+ | + | + | + | − | +12 |
| 7 | MDκ+ | + | + | + | + | +12 |
| 8 | MDκ+ | + | + | + | + | +12 |
| 9 | MDκ+ | + | + | − | − | +12 |
| 10 | MDκ+ | + | + | + | − | +12 |
| 11 | MDκ+ | + | + | + | − | del 13(q12; q14) |
| 12 | MD+ wk | + | + | − | − | del 13(q12; q14) |
| 13 | MDλ+ | + | + | − | − | del 13(q12; q22) |
| 14 | MDκ+ wk | + | + | − | − | t(13; 18)(q14; q22) |
| 15 | Mλ+ | + | + | + | − | t(13; 17)(q14; q11.2) |
| 16 | MDκ+ wk | + | + | − | − | t(13; 13)(q14; q22) |
| 17 | MDλ+ wk | + | + | + | − | del 13 (q14; q22) |
| 18 | Mλ+ wk | + | + | − | − | t(9; 13)(p11; q14) |
| 19 | Dκ+ wk | + | + | + | − | +12, del 13 (q12; q14) |
| 20 | MDκ+ | + | + | − | − | +12, t(12; 13)(q13; q14) |
| 21 | M+ wk | + | + | − | − | +12, del 13(q12; 14) |
| 22 | Mλ+ wk | + | + | + | − | +12, t(1; 13) |
Abbreviation: wk, weak.
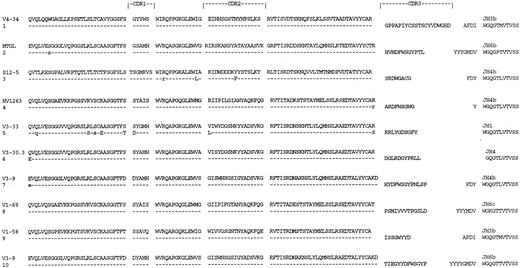
Deduced amino acid sequences of VH regions of tumor-derived clones from cases of CLL with a trisomy 12 chromosomal abnormality. Comparisons are made to the closest germ line VH genes. Uppercase, replacement mutations; lowercase, silent mutations.
Deduced amino acid sequences of VH regions of tumor-derived clones from cases of CLL with a trisomy 12 chromosomal abnormality. Comparisons are made to the closest germ line VH genes. Uppercase, replacement mutations; lowercase, silent mutations.
Deduced amino acid sequences of VH regions of tumor-derived clones from cases of CLL with a 13q14 chromosomal abnormality. Comparisons are made to the closest germ line VH genes. Uppercase, replacement mutations; lowercase, silent mutations. Replacement mutations in JH are underlined.
Deduced amino acid sequences of VH regions of tumor-derived clones from cases of CLL with a 13q14 chromosomal abnormality. Comparisons are made to the closest germ line VH genes. Uppercase, replacement mutations; lowercase, silent mutations. Replacement mutations in JH are underlined.
Deduced amino acid sequences of VH regions of tumor-derived clones from cases of CLL with both trisomy 12 and 13q14 chromosomal abnormalities. Comparisons are made to the closest germ line VH genes. Uppercase, replacement mutations; lowercase, silent mutations. Replacement mutations in JH are underlined.
Deduced amino acid sequences of VH regions of tumor-derived clones from cases of CLL with both trisomy 12 and 13q14 chromosomal abnormalities. Comparisons are made to the closest germ line VH genes. Uppercase, replacement mutations; lowercase, silent mutations. Replacement mutations in JH are underlined.
MATERIALS AND METHODS
Patients.Patients from the Haematology Clinic were staged according to the Binet classification and were previously untreated. Lymphocyte morphology was assessed at presentation on peripheral blood films stained with Grünwald Giemsa. Atypical morphology was defined as greater than 10% prolymphocytes or greater than 15% cleaved or lymphoplasmacytoid cells in the blood of patients whose predominant cell type was a small lymphocyte with coarsely clumped chromatin. All cases had monotypic expression of weak surface Ig, together with CD5 and CD23 (Tables 1 and 2).
Cytogenetic studies.Whole peripheral blood was cultured at 37°C in the presence of tetradecanoyl phorbol 12-myristate 13-acetate (TPA) (0.05 μg/mL) for 3 to 5 days. Cells were treated with colcemid (0.1 μg/mL) for 60 minutes at 37°C before harvesting. Following hypotonic treatment (KCl 0.075 mol/L for 10 minutes at 37°C), cells were resuspended in fixative (methanol:glacial acetic acid 3:1). Standard cytogenetic preparations were made, G-banded, and karyotyped according to the International System for Cytogenetic Nomenclature (1995). FISH with a biotinylated chromosome 12–specific α-satellite DNA probe (Vysis, Downers Grove, IL) to detect the number of chromosome 12 centromeres in each cell was performed in fresh material as described previously.6 Fixed TPA-stimulated cultured lymphocytes were hybridized overnight, and the number of signals in each of 200 interphase nuclei was assessed for each patient.
Microsatellite analysis.Loss of heterozygosity (LOH) was assessed at seven loci on chromosome 13 band q14.3 from the most centromeric, RB1,18 D13S153,19 D13S273,19 D13S272,19 D13S319,20 and D13S25,21 to the most telomeric, D13S262.19 Tumor and constitutional DNA was extracted from lymphocyte and neutrophil fractions by standard techniques.22 Annealing temperatures were as specified in the respective references or as determined empirically. Microsatellites D13S153, D13S273, D13S272, D13S319, and D13S262 were analyzed by incorporation of α32P during the polymerase chain reaction (PCR) and resolution on 6% polyacrylamide gel electrophoresis followed by autoradiography. The remaining PCR loci were resolved by restriction enzyme digestion and agarose gel electrophoresis. Results from tumor and constitutional fractions were compared for LOH.
Preparation of cDNA and DNA.The preferred source material was RNA, since this reduces the likelihood of amplifying an aberrantly rearranged VH gene23; cDNA was synthesized by reverse transcription using an oligo(dT) primer as previously described.23 For two of 22 cases, RNA was not available and therefore genomic DNA was extracted using the QIAamp blood kit (Qiagen, Hilden, Germany).
Amplification of VH genes.One fifth to one third of a sample of cDNA or one twentieth of a sample of genomic DNA was amplified by PCR using a mixture of oligonucleotide 5′ primers specific for each leader sequence of the VH1 to VH6 families24 together with mixed 3′ primers complementary to the germ line JH regions.25 One case (patient no. 9) failed to amplify with this combination, and an alternative mixture of 5′ primers specific for framework region 1 of VH1 to VH625 was substituted. In our hands, the VH1 leader primer also amplifies sequences from the closely related VH7 family. In all cases, PCR was performed in a final volume of 50 μL with 20 pmol of each primer, 50 μmol dNTPs, and 2.5 U Taq DNA polymerase with reaction buffer (Boehringer, Lewes, E Sussex, UK). Amplification consisted of an initial denaturation step of 5 minutes at 94°C followed by 35 cycles of 94°C, 65°C, and 72°C for 1 minute each, with a final extension step of 5 minutes at 72°C. For each PCR, a control with no added template was used to check for contamination.
Cloning and sequencing of PCR products.Cloning of gel-purified products into pGEM-TA vector was performed.23,25 Following transformation of JM109 competent cells, clones found to contain an insert of appropriate size by restriction analysis of plasmid DNA were sequenced.23,25 A minimum of five bacterial colonies were analyzed. Nucleotide sequences were aligned to EMBL/GenBank and current databases (V-BASE sequence directory,10 using MacVector 4.0 sequence analysis software; International Biotechnologies Inc, New Haven, CT).
RESULTS
Phenotypic and karyotypic analysis.All cases expressed CD5 and CD23 as defining features (Table 2). Surface Ig was low, with a tendency to be slightly higher in cases with the trisomy 12 abnormality, as previously described.8 Patients no. 1 to 10 (Table 2) with trisomy 12 all had cells with atypical morphology and had more frequent expression of CD22 and FMC7. Of these 10 cases, eight had trisomy 12 as a sole abnormality, and none had evidence of loss of 13q14 by microsatellite analysis. Patients no. 11 to 18 (Table 2) with either deletions or translocations of 13q14 had cells with typical morphology and had no detectable expression of FMC7. All eight of these cases had no other chromosomal abnormality, and there was no evidence of trisomy 12 by FISH. The four cases with both trisomy 12 and deletions or translocations of 13q14 represent a rare subset of CLL reported previously.26 In each case, interphase FISH showed that trisomy 12 and the 13q14 abnormality occurred both independently in separate cells and together within the same cell.
VH gene sequences.The deduced amino acid sequences of VH genes obtained from patients with the trisomy 12 abnormality or 13q14 abnormality are shown in Figs 1 and 2, respectively. Sequences from cases with both abnormalities are shown in Fig 3. Nucleotide sequences have been submitted to the EMBL/GenBank database (accession no. Z31382, Z31394, and Z80836-80855). In each case, five to 12 bacterial colonies were analyzed, and a clear predominant repeated sequence with the same “clonal signature” sequence in CDR3 in at least 60% of the total VH sequences was identified. Other sequences obtained had different individual CDR3 sequences characteristic of contaminating normal B cells. Since the operation of the VH primer mix has been extensively tested for bias on normal tissue and the approach has been validated by comparing identified tumor sequences with those in tumor-derived hybridomas,25 it can be concluded that the repeated sequences are consistent with a tumor population.25 The repeated sequences from each patient's tumor were identical, with only minor differences (<one of 5,000 bases) due to PCR error. This indicates that there is no intraclonal mutational heterogeneity in any of the cases. For all three subsets, a range of VH gene families was used, with no obvious differences in incidence between the groups. Overall, there was an increased usage of the VH1 family (9 of 22 cases, 45%) above that expected from the available repertoire (22%),10 and although it is not statistically significant in this small series, it supports previous observations in CLL.11 No sequences involving VH5, VH6, or VH7 families were found. A wide range of D-segment genes was used, with evidence for N-region addition in all cases (Table 3), giving rise to CDR3 sequences of variable length. There was overall predominance of the JH4 gene (10 of 22 cases), a feature found previously in both CLL and normal lymphocytes.17
Analysis of N-Region and D-Segment Genes of Cases With Trisomy 12 Only and Cases With 13q14 Abnormalities
| Patient No. . | VH Gene . | N Region . | D Region . | N Region . | JH . |
|---|---|---|---|---|---|
| Trisomy 12 | |||||
| VH4-34 | D4 | JH3 | |||
| AGA | TATTGTAGTAGTACCAGCTGCTATGCC | GCT | |||
| 1 | --- | GGACCACCTGCGGATT | -------------------------T- | GATTGGGGGCATGATGC | --- |
| MTGL | DXP4 | JH6 | |||
| AGA | ACGATTTTTGGAGTGGTT | TAC | |||
| 2 | --- | CATGTTA | ------------------ | ACCCAACGCTC | --- |
| S12-5 | DHQ52 | JH4b | |||
| CGG | ACTGGGG | CTT | |||
| 3 | --- | AGTAGAG | ------- | GGCTTGTGG | --- |
| HV1263 | DXP4 | JH4b | |||
| GAG | GATTTTTGGAGTGGTTGG | CTA | |||
| 4 | --- | AGCGAGG | ------------------ | GG | --- |
| DP-50 | DK1D3 | JH1 | |||
| GAG | CGTAGCCACTACGGTGATT | ACT | |||
| 5 | --- | GT | ---C---T--------C- | CAGGCTTCT | --- |
| DP-46 | DK4D1 | JH4 | |||
| CGA | CAGCTATGG ATACACCA | GGC | |||
| 6 | --- | AGATGGCCT | ---AG---- ----C--- | GGCTACTG | --- |
| V3-9 | DXP4D21-7RC | JH4b | |||
| GAT | ATTACGATTTTTGGAGTGGTTA CCCGAACATA | TTT | |||
| 7 | --- | C | ---------------------- -------C-- | TCTCCA | --- |
| DP-10 | D21-9 | JH6 | |||
| ACT | ACTATGATAGTAGTGGTTA | TCC | |||
| 8 | --- | CCC | -G----------------- | CCCCGGGA | --- |
| V1-58 | DN1 | JH3b | |||
| GCG | AGCAGCAGCTGGTAC | GTA | |||
| 9 | --- | ATA | -----TG-------- | TATG | --- |
| V3-9 | DXP4 | JH6b | |||
| ATA | TATTACGATTTTTGGAGTGGTT | CCT | |||
| 10 | --- | CAATTGAGGGT | ---------------------- | ACC | --- |
| V1-69 | D3 | JH3b | |||
| AGA | TAGCAATCACC | GTG | |||
| 11 | --- | GTATCCAGAGGG | ------C--A- | GAC | --- |
| V1-2 | DA5 | JH4b | |||
| GAG | GACTATGGTG | GAC | |||
| 12 | A-- | GCTGGGAGGG | -----C---- | ACTTTAACCCCAGTT | --- |
| 3d21 | DM1 | JH4b | |||
| GAG | GGAACTAC | ACT | |||
| 13 | --- | GGATAGTG | -------- | TGACGGC | --- |
| V1-3 | DXP′1 | JH4b | |||
| GAG | TGGTTCGGGG | GCT | |||
| 14 | --- | GGGCAGTGGC | -------T-- | G | --- |
| V1-2 | DN1 | JH4b | |||
| AGA | GCAGGCAG | CCT | |||
| 15 | --- | TCTCGTCTCAG | ---T--- | GTC | --- |
| V3-23 | DXP′1 | JH4b | |||
| AGA | CTATGGTTCGGGGAGTTAT | TTT | |||
| 16 | --- | CCT | --------------A---- | CGT | --- |
| V4-34 | D2 | JH6b | |||
| GAG | TATTGTAGTGGTGGTAGCTGCTACTCC | CAG | |||
| 17 | --- | CCGCTTT | ---------------G------C---- | CCG | --- |
| V3-48 | DM5-a | JH6b | |||
| GTG | CTGGAACA | ATG | |||
| 18 | --- | TC | -----G-- | GGCGAACGAT | --- |
| V1-2 | D4 | JH6b | |||
| GAG | GGATATTGTAGTAGTACCAGCTGCTAT | AAC | |||
| 19 | --- | CCCC | ----------T---------------- | GGAA | --- |
| V1-69 | D21-9 | JH4b | |||
| GAG | CTATGATAGTAGTGGTTATTACTA | CCT | |||
| 20 | --- | GCACAC | ------------------------ | TCTGG | --- |
| V4-34 | DXP′1 | JH5a | |||
| AGG | ACTATGGTTCGGGGAGTTATTATAA | GAC | |||
| 21 | --- | CGGGG | ------C---------C-----C-- | TCCCCTG | --- |
| V3-30 | DXP′5a | JH4b | |||
| CGA | AACCAGTCC | TTT | |||
| 22 | --- | AGGATCG | -------T- | GTGGTCT | --- |
| Patient No. . | VH Gene . | N Region . | D Region . | N Region . | JH . |
|---|---|---|---|---|---|
| Trisomy 12 | |||||
| VH4-34 | D4 | JH3 | |||
| AGA | TATTGTAGTAGTACCAGCTGCTATGCC | GCT | |||
| 1 | --- | GGACCACCTGCGGATT | -------------------------T- | GATTGGGGGCATGATGC | --- |
| MTGL | DXP4 | JH6 | |||
| AGA | ACGATTTTTGGAGTGGTT | TAC | |||
| 2 | --- | CATGTTA | ------------------ | ACCCAACGCTC | --- |
| S12-5 | DHQ52 | JH4b | |||
| CGG | ACTGGGG | CTT | |||
| 3 | --- | AGTAGAG | ------- | GGCTTGTGG | --- |
| HV1263 | DXP4 | JH4b | |||
| GAG | GATTTTTGGAGTGGTTGG | CTA | |||
| 4 | --- | AGCGAGG | ------------------ | GG | --- |
| DP-50 | DK1D3 | JH1 | |||
| GAG | CGTAGCCACTACGGTGATT | ACT | |||
| 5 | --- | GT | ---C---T--------C- | CAGGCTTCT | --- |
| DP-46 | DK4D1 | JH4 | |||
| CGA | CAGCTATGG ATACACCA | GGC | |||
| 6 | --- | AGATGGCCT | ---AG---- ----C--- | GGCTACTG | --- |
| V3-9 | DXP4D21-7RC | JH4b | |||
| GAT | ATTACGATTTTTGGAGTGGTTA CCCGAACATA | TTT | |||
| 7 | --- | C | ---------------------- -------C-- | TCTCCA | --- |
| DP-10 | D21-9 | JH6 | |||
| ACT | ACTATGATAGTAGTGGTTA | TCC | |||
| 8 | --- | CCC | -G----------------- | CCCCGGGA | --- |
| V1-58 | DN1 | JH3b | |||
| GCG | AGCAGCAGCTGGTAC | GTA | |||
| 9 | --- | ATA | -----TG-------- | TATG | --- |
| V3-9 | DXP4 | JH6b | |||
| ATA | TATTACGATTTTTGGAGTGGTT | CCT | |||
| 10 | --- | CAATTGAGGGT | ---------------------- | ACC | --- |
| V1-69 | D3 | JH3b | |||
| AGA | TAGCAATCACC | GTG | |||
| 11 | --- | GTATCCAGAGGG | ------C--A- | GAC | --- |
| V1-2 | DA5 | JH4b | |||
| GAG | GACTATGGTG | GAC | |||
| 12 | A-- | GCTGGGAGGG | -----C---- | ACTTTAACCCCAGTT | --- |
| 3d21 | DM1 | JH4b | |||
| GAG | GGAACTAC | ACT | |||
| 13 | --- | GGATAGTG | -------- | TGACGGC | --- |
| V1-3 | DXP′1 | JH4b | |||
| GAG | TGGTTCGGGG | GCT | |||
| 14 | --- | GGGCAGTGGC | -------T-- | G | --- |
| V1-2 | DN1 | JH4b | |||
| AGA | GCAGGCAG | CCT | |||
| 15 | --- | TCTCGTCTCAG | ---T--- | GTC | --- |
| V3-23 | DXP′1 | JH4b | |||
| AGA | CTATGGTTCGGGGAGTTAT | TTT | |||
| 16 | --- | CCT | --------------A---- | CGT | --- |
| V4-34 | D2 | JH6b | |||
| GAG | TATTGTAGTGGTGGTAGCTGCTACTCC | CAG | |||
| 17 | --- | CCGCTTT | ---------------G------C---- | CCG | --- |
| V3-48 | DM5-a | JH6b | |||
| GTG | CTGGAACA | ATG | |||
| 18 | --- | TC | -----G-- | GGCGAACGAT | --- |
| V1-2 | D4 | JH6b | |||
| GAG | GGATATTGTAGTAGTACCAGCTGCTAT | AAC | |||
| 19 | --- | CCCC | ----------T---------------- | GGAA | --- |
| V1-69 | D21-9 | JH4b | |||
| GAG | CTATGATAGTAGTGGTTATTACTA | CCT | |||
| 20 | --- | GCACAC | ------------------------ | TCTGG | --- |
| V4-34 | DXP′1 | JH5a | |||
| AGG | ACTATGGTTCGGGGAGTTATTATAA | GAC | |||
| 21 | --- | CGGGG | ------C---------C-----C-- | TCCCCTG | --- |
| V3-30 | DXP′5a | JH4b | |||
| CGA | AACCAGTCC | TTT | |||
| 22 | --- | AGGATCG | -------T- | GTGGTCT | --- |
Mutational Frequency in VH Sequences of Cases of CD5+ CLL
| Case No. . | VH Family . | GL Donor . | Mutation (%) . | |
|---|---|---|---|---|
| . | . | . | . | . |
| Trisomy 12 | ||||
| 1 | VH4 | V4-34 | DP-63 | 0 |
| 2 | VH3 | MTGL | 0 | |
| 3 | VH2 | S12-5 | 0.7 | |
| 4 | VH1 | HV1263 | 0 | |
| 5 | VH3 | V3-33 | DP-50 | 2.7 |
| 6 | VH3 | V3-30 | DP-46 | 0 |
| 7 | VH3 | V3-9 | DP-31 | 0 |
| 8 | VH1 | V1-69 | DP-10 | 0 |
| 9 | VH1 | V1-58 | DP-2 | 0 |
| 10 | VH3 | V3-9 | DP-31 | 0 |
| 13q14 | ||||
| 11 | VH1 | V1-69 | DP-10 | 7.1 |
| 12 | VH1 | V1-2 | DP-75 | 7.5 |
| 13 | VH3 | V3-30.3 | DP-46 | 4.8 |
| 14 | VH1 | V1-3 | DP-25 | 1.4 |
| 15 | VH1 | V1-2 | DP-8 | 8.0 |
| 16 | VH3 | V3-23 | DP-47 | 8.2 |
| 17 | VH4 | V4-34 | DP-63 | 6.2 |
| 18 | VH3 | V3-48 | DP-51 | 3.8 |
| Trisomy 12 and 13q14 | ||||
| 19 | VH1 | V1-2 | DP-8 | 0 |
| 20 | VH1 | V1-69 | DP-10 | 0 |
| 21 | VH4 | V4-34 | DP-63 | 5.5 |
| 22 | VH3 | V3-30 | DP-46 | 10.0 |
| Case No. . | VH Family . | GL Donor . | Mutation (%) . | |
|---|---|---|---|---|
| . | . | . | . | . |
| Trisomy 12 | ||||
| 1 | VH4 | V4-34 | DP-63 | 0 |
| 2 | VH3 | MTGL | 0 | |
| 3 | VH2 | S12-5 | 0.7 | |
| 4 | VH1 | HV1263 | 0 | |
| 5 | VH3 | V3-33 | DP-50 | 2.7 |
| 6 | VH3 | V3-30 | DP-46 | 0 |
| 7 | VH3 | V3-9 | DP-31 | 0 |
| 8 | VH1 | V1-69 | DP-10 | 0 |
| 9 | VH1 | V1-58 | DP-2 | 0 |
| 10 | VH3 | V3-9 | DP-31 | 0 |
| 13q14 | ||||
| 11 | VH1 | V1-69 | DP-10 | 7.1 |
| 12 | VH1 | V1-2 | DP-75 | 7.5 |
| 13 | VH3 | V3-30.3 | DP-46 | 4.8 |
| 14 | VH1 | V1-3 | DP-25 | 1.4 |
| 15 | VH1 | V1-2 | DP-8 | 8.0 |
| 16 | VH3 | V3-23 | DP-47 | 8.2 |
| 17 | VH4 | V4-34 | DP-63 | 6.2 |
| 18 | VH3 | V3-48 | DP-51 | 3.8 |
| Trisomy 12 and 13q14 | ||||
| 19 | VH1 | V1-2 | DP-8 | 0 |
| 20 | VH1 | V1-69 | DP-10 | 0 |
| 21 | VH4 | V4-34 | DP-63 | 5.5 |
| 22 | VH3 | V3-30 | DP-46 | 10.0 |
Analysis of the Distribution of Somatic Mutations
| Patient No. . | Germ Line . | CDR/FWR . | Calculated R:S . | Observed . | P* . | ||
|---|---|---|---|---|---|---|---|
| . | . | . | . | R . | S . | R:S . | . |
| 11 | V1-69 | CDR | 3.4 | 5 | 3 | 1.7 | .16 |
| FWR | 3.0 | 8 | 5 | 1.6 | .04 | ||
| 12 | V1-2 | CDR | 4.3 | 7 | 0 | ∞ | .06 |
| FWR | 3.0 | 8 | 7 | 1.1 | .02 | ||
| 13 | V3-30.3 | CDR | 3.9 | 8 | 0 | ∞ | <.01 |
| FWR | 2.9 | 4 | 2 | 2.0 | .02 | ||
| 14 | V1-3 | CDR | 4.3 | 0 | 0 | ∞ | .45 |
| FWR | 3.0 | 3 | 1 | 3.0 | .33 | ||
| 15 | V1-2 | CDR | 4.3 | 4 | 3 | 1.3 | .21 |
| FWR | 3.0 | 12 | 4 | 3.0 | .14 | ||
| 16 | V3-23 | CDR | 3.5 | 8 | 2 | 4.0 | .03 |
| FWR | 2.9 | 6 | 8 | 0.8 | <.01 | ||
| 17 | V4-34 | CDR | 4.4 | 3 | 4 | 0.8 | .24 |
| FWR | 2.8 | 8 | 3 | 2.7 | .10 | ||
| 18 | V3-48 | CDR | 4.7 | 4 | 0 | ∞ | .10 |
| FWR | 2.8 | 5 | 2 | 2.5 | .18 | ||
| 21 | V4-34 | CDR | 4.4 | 3 | 0 | ∞ | .25 |
| FWR | 2.8 | 11 | 2 | 5.5 | .14 | ||
| 22 | V3-30 | CDR | 3.9 | 15 | 1 | 15.0 | <.01 |
| FWR | 2.9 | 9 | 4 | 2.3 | .01 | ||
| Patient No. . | Germ Line . | CDR/FWR . | Calculated R:S . | Observed . | P* . | ||
|---|---|---|---|---|---|---|---|
| . | . | . | . | R . | S . | R:S . | . |
| 11 | V1-69 | CDR | 3.4 | 5 | 3 | 1.7 | .16 |
| FWR | 3.0 | 8 | 5 | 1.6 | .04 | ||
| 12 | V1-2 | CDR | 4.3 | 7 | 0 | ∞ | .06 |
| FWR | 3.0 | 8 | 7 | 1.1 | .02 | ||
| 13 | V3-30.3 | CDR | 3.9 | 8 | 0 | ∞ | <.01 |
| FWR | 2.9 | 4 | 2 | 2.0 | .02 | ||
| 14 | V1-3 | CDR | 4.3 | 0 | 0 | ∞ | .45 |
| FWR | 3.0 | 3 | 1 | 3.0 | .33 | ||
| 15 | V1-2 | CDR | 4.3 | 4 | 3 | 1.3 | .21 |
| FWR | 3.0 | 12 | 4 | 3.0 | .14 | ||
| 16 | V3-23 | CDR | 3.5 | 8 | 2 | 4.0 | .03 |
| FWR | 2.9 | 6 | 8 | 0.8 | <.01 | ||
| 17 | V4-34 | CDR | 4.4 | 3 | 4 | 0.8 | .24 |
| FWR | 2.8 | 8 | 3 | 2.7 | .10 | ||
| 18 | V3-48 | CDR | 4.7 | 4 | 0 | ∞ | .10 |
| FWR | 2.8 | 5 | 2 | 2.5 | .18 | ||
| 21 | V4-34 | CDR | 4.4 | 3 | 0 | ∞ | .25 |
| FWR | 2.8 | 11 | 2 | 5.5 | .14 | ||
| 22 | V3-30 | CDR | 3.9 | 15 | 1 | 15.0 | <.01 |
| FWR | 2.9 | 9 | 4 | 2.3 | .01 | ||
Values in boldface type reach significance (P < .05). Underlined values indicate significant clustering in CDRs.
Abbreviation: P, probability of obtaining the number of R mutations found in CDRs by chance.
Analysis of somatic mutation.Comparison to the closest germ line genes in the database showed marked differences in the degree of somatic mutation between the three subsets (Figs 1-3). In cases with trisomy 12, a low level was observed, with eight of 10 cases being identical to the germ line sequence (Table 4 and Fig 1). VH sequences of patients with the 13q14 abnormality (Fig 2) showed a greater degree of somatic mutation ranging from 1.4% to 8.2% (mean ± SD, 6.5% ± 1.67%; Table 4). Differences in the incidence of mutational events (mean percent mutation frequency) between the two groups were highly statistically significant (P < .0003 by Mann-Whitney U test). Cases with both chromosomal abnormalities showed a mixed pattern, with two of four being identical to the germ line sequence and two of four having significant levels of somatic mutation (Table 4 and Fig 3). Analysis of the distribution of replacement and silent mutations in the mutated sequences (Table 5) using the method of Chang and Casali27 showed that three of 10 had significant clustering of replacement amino acids in CDR1 and CDR2 indicative of antigen selection. In these three cases and another two cases, there was evidence in FWRs for conservation of sequence (Table 5).
DISCUSSION
Although CLL is the most common leukemia in the West and has been studied extensively, the cell of origin remains undefined (reviewed in Caligaris-Cappio et al28 ). There is no unequivocal normal B-cell counterpart with the characteristic phenotypic profile of CLL cells expressing CD5 and CD23 together with low levels of surface IgM and IgD. One candidate is the long-lived, recirculating subpopulation of mantle zone (MZ) cells.28 However, although a proportion of MZ cells express CD23, they differ from CLL in expressing high levels of surface Ig.28,29 One suggestion that has been made is that the low surface Ig of CLL cells may be due to induction of anergy by exposure to self-antigens.29
Since V genes undergo rearrangements and somatic mutations during B-cell differentiation, V genes of B-cell tumors contain information that might relate to the clonal history of the cell of origin. Studies of VH gene usage in B-cell tumors can reveal asymmetric use of VH genes from the available repertoire. In CLL, there is an apparent increased usage of the VH1-69 (51p1) gene from the VH1 family.11 This is supported by our study of 22 cases defined by chromosomal abnormalities, with no obvious difference in VH gene usage between the subsets. Although the current view is that CLL arises from cells that contain unmutated V genes,29 there are many CLL-derived VH sequences that deviate from the germ line sequence by greater than 2%.17 In our study of CD5+CD23+ CLL, cases with trisomy 12 appear to reflect the current view in having few or no somatic mutations. In fact, we can now add two of two further cases with this pattern (Dr G. Dighiero, personal communication, December 1996). The cells could relate to the normal IgM+IgD+CD23+ cells of the MZ, which are apparently unmutated.30 It could be concluded that this subset of CLL is more likely to accumulate the trisomy 12 abnormality in some members of the clone.
In contrast, the category with 13q14 abnormalities, although using a similar range of genes, contains a considerable number of somatic mutations. Since the mutation machinery is thought to be activated only after B cells reach the germinal center at the centroblast stage,14,15 the cell of origin is likely to have traversed this site. In tonsil, this transition is associated with loss of IgD and CD23 expression,15,31 which has clearly not occurred in the CLL cells. It is certainly known that somatic mutations can be accumulated before isotype switch14-17 and that a proportion of blood B cells carry mutated VH genes, but these cells are thought to express only IgM.32 In fact, a recent analysis of tonsillar B lymphocytes also found that IgD+ cells do not carry somatic mutations and are therefore unlikely to be memory B cells.33 However, there is evidence in a proportion of tonsils for the existence of a population of CD70+IgD+ germinal center cells that may represent recent immigrants in the process of forming a germinal center.34 Also, mature IgD-expressing B cells have been detected in bone marrow, and it has been suggested that these cells may be memory cells that have been selected in the periphery for re-entry into this site.35 The status of the V genes of either of these B-cell populations is not yet known, but if they have been exposed to the mutation mechanism, they could be candidates for the normal counterparts of the subset of CLL that is defined by the 13q14 abnormality.
The somatically mutated CLL cases therefore may have arisen from B cells that have undergone mutational events but do not isotype-switch, or lose expression of IgD, before exit to the blood. Interestingly, a cell line from a case of Burkitt's lymphoma (ELI) that was IgM+IgD+ also had a high degree of somatic mutation,36 supporting the fact that among B-cell tumors expression of IgM plus IgD can be accompanied by somatic mutation. In three of 10 CLL cases with mutations, the pattern was consistent with a role for antigen selection, supporting a memory function.14-16 The fact that there was no intraclonal heterogeneity suggests that, in contrast to follicular lymphoma and Burkitt's lymphoma, the tumor cells are no longer influenced by the mutational mechanism associated with the germinal center.36-38 However, V genes in tumor cells in CLL may not be as static as has been deduced from phenotypic analysis, since transcripts indicative of isotype-switch events to Cγ and Cα have been observed in typical IgM+ CLL.39-41 Although it could be argued that any of these features are due to neoplastic changes, they may in fact reveal aspects of normal B-cell behavior.
Chromosomal abnormalities are not found in all members of the neoplastic clone,26 and are therefore unlikely to be initiating events. For CLL, the transformation event is unknown, but our results indicate that there may be two subsets of CLL that transform at distinct stages of differentiation and that each go on to accumulate different chromosomal abnormalities. The subset that remains unmutated, and that may derive from naive B cells, appears prone to accumulate the trisomy 12 abnormality. In contrast, the mutated subset, perhaps derived from more mature B cells, appears more prone to abnormalities of 13q14. In the rare subset of CLL with both chromosomal abnormalities, the mutational pattern is heterogeneous. These findings reveal heterogeneity in CLL indicative of differences in the cell of origin that could relate to pathogenesis and disease progression. They also suggest that chromosomal changes in CLL, which are likely to be subsequent to the initiating transformation event, may be characteristic of the identified subsets.
ACKNOWLEDGMENT
We thank A. Gardiner, S. Mould, and R. Ibbotson for performing the cytogenetic analyses.
Supported by Tenovus, the Kay Kendall Leukaemia Fund, and the Cancer Research Campaign, UK.
Address reprint requests to Freda K. Stevenson, DPhil, Molecular Immunology Group, Tenovus Laboratory, Southampton University Hospitals, Tremona Rd, Southampton SO16 6YD, UK.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal