Abstract
The recurrent translocation t(11; 16)(q23; p13) has been reported to be associated with therapy-related acute leukemia. The MLL gene involved in other 11q23 abnormalities was also rearranged by this translocation. We analyzed two patients with myelodysplastic syndrome with t(11; 16) and showed that the MLL gene on 11q23 was fused with CREB-binding protein (CBP) gene on 16p13 in these patients. The CBP gene encodes a transcriptional adaptor/coactivator protein and it is mutated in patients with Rubinstein-Taybi syndrome. The CBP gene is also involved in acute myeloid leukemia (AML) with t(8; 16)(p11; p13). In-frame MLL-CBP fusion transcripts combine the MLL AT-hook motifs and DNA methyltransferase homology region with a largely intact CBP. Our results combined with the finding of the MOZ-CBP fusion in t(8; 16)-AML suggest that the CBP gene may be associated with leukemogenesis through translocations.
CHROMOSOMAL translocations are the most common genetic events observed in hematopoietic malignancies.1,2 It has been thought that specific genes are associated with specific translocations.3 However, recent molecular and cytogenetic studies have shown that some genes involved in specific translocations are also associated with other translocations, such as the AML1 gene in t(8; 21)-acute myeloid leukemia (AML)4 and t(12; 21)-acute lymphoblastic leukemia (ALL),5 and the NPM gene in t(2; 5)–non-Hodgkin's lymphoma (NHL)6 and the t(3; 5)-myelodysplastic syndrome (MDS),7 and that some tumor-suppressor genes are occasionally associated with chromosomal translocations.8,9 The MLL gene10 (also called ALL-1, HRX, and Htrx-1) has been identified in 11q23 translocations,11-13 and its rearrangement was found in the majority of infant14-16 and secondary leukemias.17,18 This gene forms fusion transcripts with over 10 partner genes.11,12,19,20 However, it remains to be elucidated whether the MLL gene, the partner genes, or the fusion transcripts play a critical role in leukemogenesis. The CREB-binding protein (CBP) gene, encoding a transcriptional adaptor/coactivator protein, resides on 16p1321 and it is mutated in patients with Rubinstein-Taybi syndrome.22 Recently, the CBP gene was found to be translocated in AML with t(8; 16)(p11; p13).23,24 The recurrent translocation t(11; 16) (q23; p13) is associated with therapy-related acute leukemia.17 25 Here we identified that the MLL gene was fused with CBP gene in two patients of MDS with t(11; 16). Our results combined with the finding of the MOZ-CBP fusion in t(8; 16)-AML suggest that the CBP gene may be associated with leukemogenesis through translocations.
MATERIALS AND METHODS
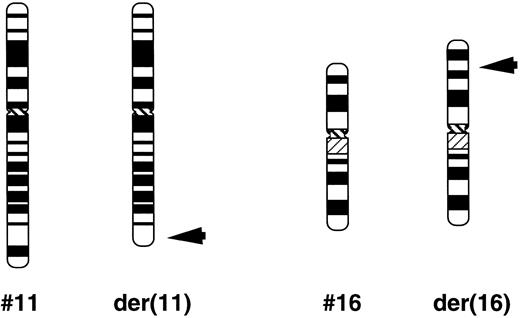
Patients.Patient 1, a 2-year-old girl, was diagnosed as refractory anemia with excess of blasts in transformation (RAEB-T). Patient 2, a 6-year-old boy, was initially diagnosed as AML(M2) with t(8; 21)(q22; q22). He achieved a complete remission by chemotherapy including etoposide. After 14 months he developed RAEB-T. Patient samples were obtained at diagnosis from bone marrow (BM) aspirations of both patients with RAEB-T, and their karyotypes showed t(11; 16)(q23; p13) (Fig 1).
Schematic partial karyotype of t(11; 16) (q23; p13). The normal chromosomes of nos. 11 and 16 are on the left in each pair. Arrowheads indicate the breakpoints in the translocation chromosome on the right.
Schematic partial karyotype of t(11; 16) (q23; p13). The normal chromosomes of nos. 11 and 16 are on the left in each pair. Arrowheads indicate the breakpoints in the translocation chromosome on the right.
Cytogenetic studies.The chromosomes of BM cells from patients 1 and 2 were analyzed by the regular trypsin-Giemsa- or Q-banding method as previously described.16 26
Southern and Northern blot analyses.High-molecular-weight DNA was extracted from BM cells from patient 1 by proteinase K digestion and phenol/chloroform extraction. Ten micrograms of DNA was digested with appropriate restriction enzymes, subjected to electrophoresis on 0.8% agarose gels, transferred to charged nylon filters (Pall BioSupport, Tokyo, Japan), and hybridized to DNA probes labeled by the random hexamer method.16 19
Poly(A)+ RNA from BM cells of patient 1 was extracted with a Fast Track mRNA Isolation Kit (Invitrogen, San Diego, CA). One microgram of poly (A)+ mRNAs was electrophoresed and transferred onto nylon filters. Blots were hybridized to 32P-labeled probes under the described conditions.19
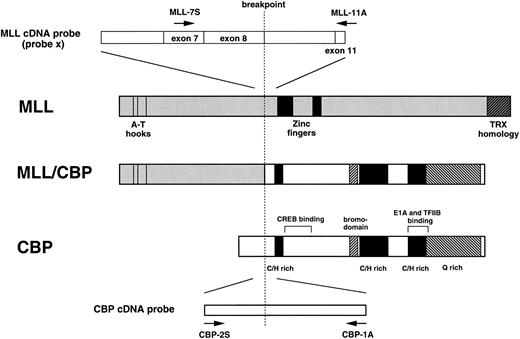
A 0.9-kb BamHI fragment (designated probe x) derived from MLL cDNA,27 and a 0.5-kb CBP cDNA (nucleotides 701-1197 from patial sequences of CBP cDNA, GenBank accession no. U47741) were used as probes (Fig 2).
Schematic representation of the MLL, CBP, and MLL-CBP fusion proteins, and MLL and CBP cDNA probes and primers (MLL-7S, MLL-11A, CBP-2S, CBP-1A). C/H rich, cysteine/histidine rich; Q rich, glutamine rich. The position of the motifs in CBP were derived from the published protein sequence. The MLL-CBP fusion protein retains the key domains from both proteins.
Schematic representation of the MLL, CBP, and MLL-CBP fusion proteins, and MLL and CBP cDNA probes and primers (MLL-7S, MLL-11A, CBP-2S, CBP-1A). C/H rich, cysteine/histidine rich; Q rich, glutamine rich. The position of the motifs in CBP were derived from the published protein sequence. The MLL-CBP fusion protein retains the key domains from both proteins.
Reverse transcriptase-polymerase chain reaction (RT-PCR).Total cellular RNA was extracted from BM cells of patients 1 and 2 by the acid guanidine isothiocyanate-phenol-chloroform method.28 Four micrograms of total RNA was reverse transcribed to cDNA in a total volume of 20 μL with random hexamers and 20 U of reverse transcriptase (AMV) (Boehringer Mannheim). One twentieth of the cDNA was amplified by PCR in a total volume of 100 μL with 50 mmol/L KCl, 1.5 mmol/L MgCl2 , 10 mmol/L Tris-HCl (pH 9.0 at room temperature), 25 pmol/L of each primer, 75 μmol/L of each dNTP, and 2.5 U of Taq polymerase (Boehringer Mannheim). After 30 rounds of PCR (30 seconds at 94°C, 30 seconds at 55°C, and 1 minute at 72°C), 5 μL of PCR product was electrophoresed in a 3% agarose gel. Primers used were as follows: MLL-7S, 5′-TCCTCAGCACTCTCTCCAAT-3′; MLL-11A, 5′-TTTGCCTGGAGTTGTGGATC-3′; CBP-2S, 5′-TGAATGGATCTCTTGGGGCT-3′; CBP-1A, 5′-GTTTGCTTGCTCTCGTCTCT-3′ (Fig 2).
Cloning and sequencing of PCR products.PCR products were cloned into the TA cloning vector (Invitrogen). Nucleotide sequences were determined by the fluorometric method (Dye Terminator Cycle Sequencing Kit; Applied Biosystems, Urayasu, Japan).
RESULTS
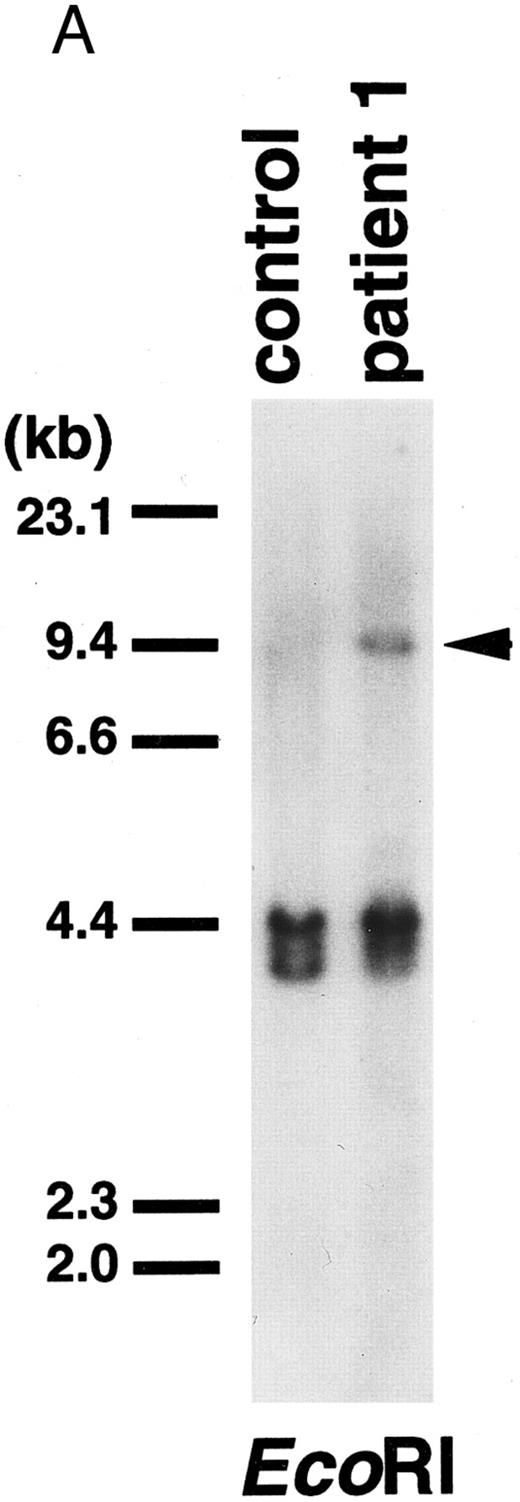
MLL and CBP genes were involved in t(11; 16)-MDS.Cells from two children with MDS were cytogenetically characterized as t(11; 16)(q23; p13) (Fig 1). We identified the chromosomal breakpoint within the breakpoint cluster region of MLL gene at 11q23 by Southern blotting using a cDNA probe (probe x) (Figs 2 and 3A) that spans exons 5 through 11 in the MLL locus.27
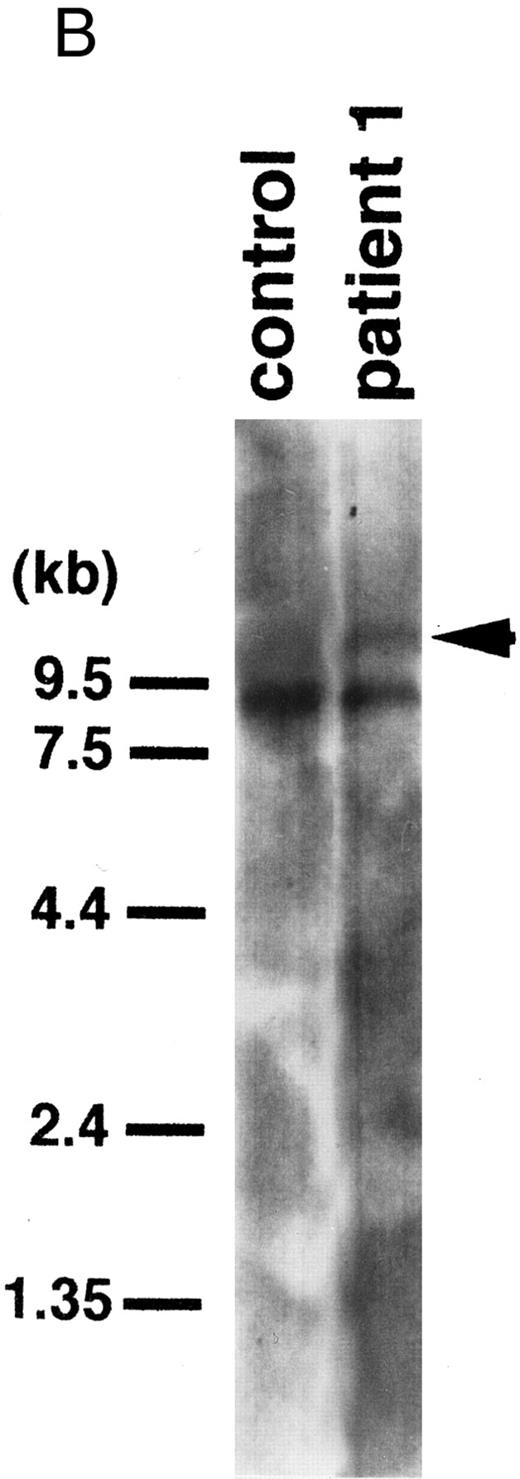
(A) Southern blot of DNA digested with EcoRI and probed with the 0.9-kb fragment of the MLL gene. Control, peripheral lymphocytes. Patient 1 exhibited a rearranged band (arrowhead) with this probe. (B) Northern blot of RNAs from patient 1 and a control leukemic cell line (BALM14) using a 0.5-kb CBP cDNA fragment as a probe. Patient 1 showed an abnormal transcript (arrowhead).
(A) Southern blot of DNA digested with EcoRI and probed with the 0.9-kb fragment of the MLL gene. Control, peripheral lymphocytes. Patient 1 exhibited a rearranged band (arrowhead) with this probe. (B) Northern blot of RNAs from patient 1 and a control leukemic cell line (BALM14) using a 0.5-kb CBP cDNA fragment as a probe. Patient 1 showed an abnormal transcript (arrowhead).
Chromosome 16 band p13 is involved in the chromosomal aberrations, inv(16)(p13q22) and t(8; 16)(p11; p13), in AML. However, the position of the breakpoint at 16p13 in inv(16) differs from that of the t(8; 16).29 To isolate a gene on chromosome 16p13 that fuses to MLL, we performed Southern and Northern blot analyses using a CBP cDNA probe (Fig 2). Blots that were used to detect MLL rearrangements and expression were rehybridized with a probe containing 0.5-kb of the 9-kb CBP cDNA. This probe contains cDNA sequences from both sides of the breakpoint within the CBP cDNA in t(8; 16)-AML and could thus potentially detect both der(11) and der(16). Southern blotting did not detect any rearrangements with CBP cDNA. To clarify the expression of the CBP transcripts, a CBP probe was examined for hybridization to poly(A)+ mRNAs extracted from cells from patient 1, which showed a normal 9-kb CBP transcript and an additional 12-kb transcript (Fig 3B).
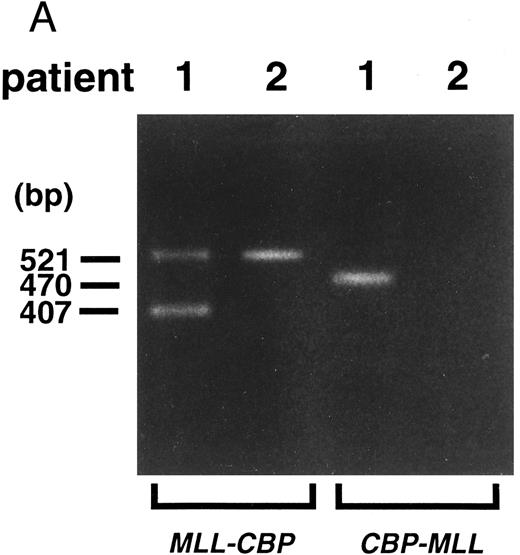
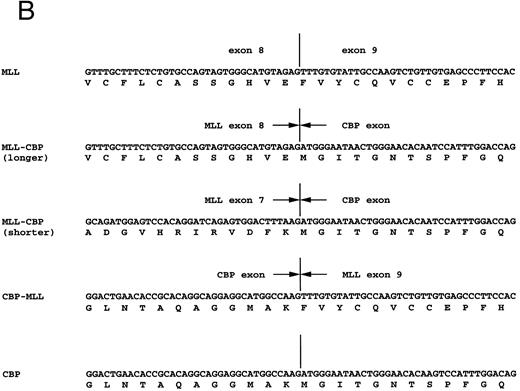
Detection of MLL-CBP fusion transcripts.Using the sense primer on MLL exon 7 (MLL-7S) and the antisense primer on CBP (CBP-1A), we obtained PCR products of 521 bp from patients 1 and 2, and of 407 bp from patient 1 (Fig 4A). Nucleotide sequencing analysis of the three amplified fragments demonstrated 197 bp of MLL exons 7 and 8 in the 5′ region, and 324 bp of CBP exon in the 3′ region of the longer fragment from both patients, and 83 bp of MLL exon 7 in the 5′ region and 324 bp of the CBP exon in the 3′ region of the shorter fragment from patient 1 (Fig 4B). These findings indicate that the two chimeric products are generated by alternative splicing at the MLL gene, which was observed in other 11q23 translocations.30 Only in patient 1 were reciprocal PCR products of CBP-MLL fusion transcripts generated by RT-PCR using the sense primer on CBP (CBP-2S) and the antisense primer on MLL exon 11 (MLL-11A) (Fig 4A and B). The fusion transcripts of MLL-CBP commonly observed were considered to play a critical role in the leukemogenesis of t(11; 16)-MDS.
(A) Identification of fusion transcripts by RT-PCR. Primers used were MLL-7S and CBP-1A (MLL-CBP), and CBP-2S and MLL-11A (CBP-MLL). (B) Partial sequences at the chimeric junctions from five cDNAs (MLL, longer and shorter fragments of MLL-CBP, CBP-MLL, and CBP). longer, longer fragment of 521 bp from patients 1 and 2; shorter, shorter fragment of 407 bp from patient 1. Vertical lines indicate the exon-exon junctions of each gene. Arrows indicate the fusion points of each cDNA.
(A) Identification of fusion transcripts by RT-PCR. Primers used were MLL-7S and CBP-1A (MLL-CBP), and CBP-2S and MLL-11A (CBP-MLL). (B) Partial sequences at the chimeric junctions from five cDNAs (MLL, longer and shorter fragments of MLL-CBP, CBP-MLL, and CBP). longer, longer fragment of 521 bp from patients 1 and 2; shorter, shorter fragment of 407 bp from patient 1. Vertical lines indicate the exon-exon junctions of each gene. Arrows indicate the fusion points of each cDNA.
DISCUSSION
The t(11; 16) has thus far been reported in two patients with therapy-related ALL and AML.17 25 Chromosomal breakpoints in 11q23 were detected within the MLL breakpoint cluster region by Southern blotting in both patients. In the present study we first identified that the MLL gene was fused to the CBP gene in two MDS-patients with t(11; 16).
The MLL gene, a human homolog of the Drosophila trithorax gene, encodes a protein of 3,972 amino acids (431 kD) containing zinc fingers and AT-hook motifs.11-13 The incidence of MLL gene rearrangements has been shown to be up to 80% of infant acute leukemia.14-16MLL gene rearrangements also occur at very high frequency in secondary acute leukemias induced by treating malignancies with inhibitors of topoisomerase II,17,18 suggesting that a reduction in the activity of topoisomerase II is one of the mechanisms for the occurrence of MLL gene rearrangement. It is noted that the MLL gene is fused with various partner genes by 11q23 chromosome translocations. Up to now, 10 partner genes for MLL have been cloned from leukemia cells with various types of reciprocal translocations, including t(4; 11), t(9; 11) and t(11; 19).11-13,19,20 They include the putative transcriptional factors (AF-4, AF-9, AF-10, AF-17, ENL),11,12,20 a target gene for Ras (AF-6),31 and an RNA polymerase II elongation factor (ELL).32 The functions of the normal MLL gene and the fusion transcripts remain unknown. Only one paper reports that chimeric mice carrying the mouse Mll-AF-9 fusion gene developed AML.33 Rearrangements of the MLL gene found in ALL, AML, and MDS suggest that this gene fused with various partner genes plays a causative role in the dysregulation of differentiation along both lymphoid and myeloid pathways.
The CBP, a family of transcriptional coactivator p300, has an adenoviral oncoprotein E1A binding domain and connects the basal transcriptional machinery to transcriptional factors such as the cyclic-adenosine monophosphate responsive element binding protein (CREB).21 CBP normally associates with p300/CBP-associated factor (P/CAF ) having intrinsic histon acetyltransferase activity.34 P/CAF, and also p300, inhibit cell-cycle progression,34 suggesting that CBP may also be involved in the regulation of cell cycle. Moreover, mutations of CBP lead to the development of Rubinstein-Taybi syndrome,22 whose patients have an increased risk of malignant tumors.35 These findings suggest that CBP may be a tumor-suppressor gene. Fusion of CBP to translocated MOZ sequences is associated with specific subtypes of AML with t(8; 16),23 24 and this leukemogenesis may be also associated with dysfunction of CBP.
A schematic representation of the predicted MLL-CBP fusion proteins is shown in Fig 2, with the predicted MLL-CBP fusion transcripts encoding a protein of 3,618 amino acids containing 1,444 amino acids from MLL (amino acids 1 to 1,444) and 2,174 amino acids from CBP (amino acids 267 to 2,440). The MLL-CBP chimeric protein retains the AT-hook domain, DNA methyltransferase homology region, and a transcriptional repression domain of MLL, and the CREB-, E1A-, TFIIB- and P/CAF-binding domains and the acetyltransferase domain36 of CBP. These findings suggest that one possible explanation for leukemogenesis of t(11; 16)-MDS is the disability of CBP to regulate the cell cycle by structural alteration through fusion with MLL. Further functional analysis of the MLL-CBP fusion gene will provide new insights into leukemogenesis by 11q23 chromosomal translocations.
ACKNOWLEDGMENT
We thank Prof Y. Igarashi (Department of Pediatrics, Hamamatsu University School of Medicine), Prof M. Yanagisawa (Department of Pediatrics, Faculty of Medicine, University of Tokyo), and I. Kitabayashi (Radiobiology Division, National Cancer Center Research Institute, Tokyo), for their advice and review. We also thank M. Seto (Laboratory of Chemotherapy, Aichi Cancer Center Research Institute) for providing the MLL cDNA probe (probe x), and S. Sohma for her technical assistance.
Supported by a Grant-in-Aid for Cancer Research from the Ministry of Health and Welfare of Japan, a Grant-in-Aid for Scientific Research on Priority Areas, and Grant-in-Aid for Scientific Research (B) and (C) from the Ministry of Education, Science, Sports and Culture of Japan.
Address reprint requests to Yasuhide Hayashi, MD, Department of Pediatrics, Faculty of Medicine, University of Tokyo, 7-3-1 Hongo Bunkyo-ku, Tokyo 113, Japan.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal