In this issue of Blood, Dong et al1 identify functional hematopoietic stem cell (HSC) clones with robust, high serially transplantable lymphomyeloid blood output and reveal the gene expression signature driving this activity.
Bone marrow transplantation remains the gold standard curative therapy for many blood disorders, including both benign (eg, severe combined immune deficiency syndromes and bone marrow failure syndromes) and malignant disorders.2,3 Yet a critical question remains, how do HSCs rapidly restore blood and immune system function posttransplantation? This unresolved challenge limits their therapeutic potential.
Dong et al have adopted a rigorous and comprehensive approach to evaluate murine HSC functionality through syngeneic single-stem cell serial transplantation experiments. HSC clones with robust, balanced lymphomyeloid blood output in a serial transplantation experiment (up to the third generation) were referred to as having high “transplantability.” These unique HSC clone characteristics align closely with the clinical “holy grail” for improving the success rate of bone marrow transplantation. This discovery challenges the current hypothesis that upon transplantation quick initial repopulation of blood cells occurs via the activity of a separate group of HSCs that are being complemented by highly self-renewing but slower-differentiating HSC populations that became major blood contributors over time.4,5 A similar subset of HSCs has been recently reported in nonhuman primates; however, the transplantability potential and detailed transcriptional signature of this subset were not determined.6 Hence the evidence provided by Dong et al complements previous studies5,6 by functionally showing that these HSC clones retain repopulating potential in serial transplantation experiments and provide molecular signatures for identified HSC clone types.
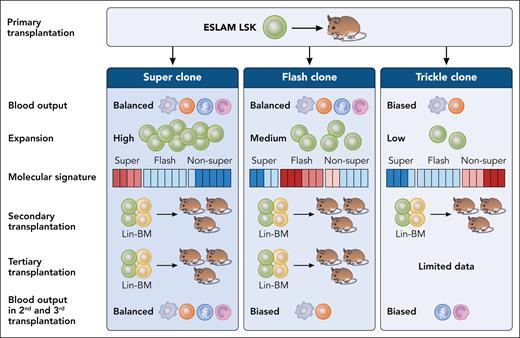
To classify HSC reconstitution patterns, the authors employed Bayesian modeling and identified 3 distinct types of clones: SUPER, FLASH, and TRICKLE (see figure). By analyzing the transcriptomic landscapes in these different clone types, Dong et al identified and validated that low expression of immunophenotypic marker Cd27 can enrich highly transplantable HSCs with a balanced lymphomyeloid output.
Upon single stem cell transplantation (Lin-Sca-1+c-Kit+CD150+48-CD201+, called ESLAM LSK) in a mouse model, HSC clones can be categorized into 3 distinct types based on their ability to produce blood cells: SUPER, FLASH and TRICKLE. These clones exhibit either balanced (multilineage) or biased (lineage restricted) differentiation potential. Each clone type demonstrates unique transplantability and contributes differently to blood production during serial transplantation. The functional properties of these HSC clones are driven by distinct molecular signatures (blue-low expression, red-high expression). Professional illustration by Patrick Lane, ScEYEnce Studios.
Upon single stem cell transplantation (Lin-Sca-1+c-Kit+CD150+48-CD201+, called ESLAM LSK) in a mouse model, HSC clones can be categorized into 3 distinct types based on their ability to produce blood cells: SUPER, FLASH and TRICKLE. These clones exhibit either balanced (multilineage) or biased (lineage restricted) differentiation potential. Each clone type demonstrates unique transplantability and contributes differently to blood production during serial transplantation. The functional properties of these HSC clones are driven by distinct molecular signatures (blue-low expression, red-high expression). Professional illustration by Patrick Lane, ScEYEnce Studios.
These novel findings have clinical implications, making it of great interest for the community to identify a human equivalent of SUPER clones. To provide a broader molecular signature, Dong et al performed an Assay for Transposase-Accessible Chromatin with sequencing (ATAC-seq) experiment in their murine system and identified an extensive list of differentially open-close chromatin regions in SUPER clones. This molecular signature could inform the search for a human equivalent of SUPER-HSC clones by using existing data sets. To refine current immunophenotypic characteristics of SUPER-HSC clones, a complementary approach incorporating index sorting during HSC isolation for primary transplantation would further expand our ability to enrich these cells. Results from these efforts could inform new recommendations for HSC transplantation to increase its success rate.
SUPER-HSC clones have a unique potential to robustly regenerate analyzed lymphomyeloid blood cells. Incorporating platelets and erythroid cells in the analysis7,8 could further strengthen the translational potential of the presented findings. These lineages are of great clinical importance due to the frequently observed slow reconstitution upon transplantation.
The authors have primarily focused on the characterization of SUPER clones. Among identified HSC clone types, there might be other HSC groups that can provide similar transplantability and multilineage blood production. The functional characteristics of FLASH and TRICKLE clones (P3 output; see Figure 3B in the article by Dong et al that begins on page 546) resemble some of the properties identified in low-output clones.9 Therefore, further investigation of these clones in serial transplantation experiments and their molecular analyses could provide new immunophenotypic and molecular markers to complement those of SUPER clones for enhanced blood and immune system restoration upon transplantation.
Dong et al have performed extensive single-cell transcriptomic analyses of HSC clones. These efforts could be expanded in the future by incorporating isoform-level information to discover novel transcripts potentially further distinguishing identified HSC clone types.
The authors have solely focused on the cell-intrinsic characterization of SUPER-HSC clones. Future studies will decode microenvironments supporting these unique SUPER-HSC clones. It would be also compelling to test whether other HSC clones can be reprogrammed into balanced, high-output SUPER clones through perturbing identified signaling pathways.
The overall study by Dong et al provides novel insights into HSC biology and function. To broaden its clinical implications, it would be important to identify the human equivalent of these SUPER-HSC clones, the niche supporting these cells, and the impact of donor source (ie, allogeneic or syngeneic) and to evaluate the potential of reprogramming other HSC clone types into SUPER clones.
Conflict-of-interest disclosure: E.E.W. declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal