In this issue of Blood, Escherich et al1 use preclinical models to demonstrate that inotuzumab ozogamicin (InO) sensitivity in B-cell acute lymphoblastic leukemia (B-ALL) depends on the stage of B-cell developmental arrest. They identified CD22—the surface antigen targeted by InO—as a direct transcriptional target of early B-cell factor 1 (EBF1) and showed that B-ALL samples exhibiting gene-regulatory profiles, resembling those of normal Pre-pro-B cells at the onset of EBF1 activation, had reduced responsiveness to InO.
Successes achieved over the last decades in B-ALL therapy have been built on the individualized adjustment of chemotherapy intensity and stratified use of allogeneic stem cell transplantation guided by biological baseline characteristics and the clearance of measurable residual disease. Immunotherapies such as InO, blinatumomab (Blina), and rituximab are reshaping this landscape by shifting from salvage settings to frontline therapy. Up-front use of InO or Blina showed remarkable efficacy in older patients2 and infants,3 both more vulnerable to chemotherapy-associated toxicities, and Blina was recently approved for frontline therapy of adult patients in molecular remission.4 A shared dependency among these immunotherapies is the sustained expression of lineage-specific target antigens such as CD22, CD20, or CD19. Loss of target antigen expression either through acquisition of deleterious genomic variants,5 downregulation, or complete switch of hematopoietic lineage are modes of resistance to these treatments. A better understanding of the underlying mechanisms is necessary to adapt up-front treatment allocation.
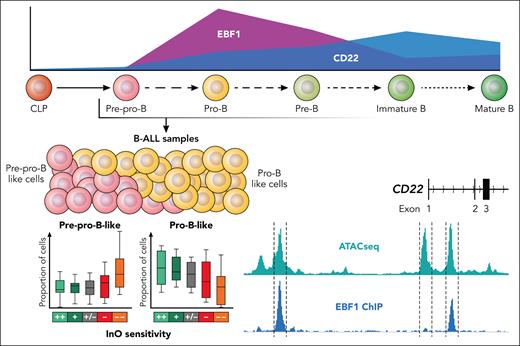
The membrane glycoprotein CD22 is an attractive immunotherapy target due to its specificity to the B lineage and its rapid internalization after ligand binding—a feature particularly amenable to antibody–drug conjugate strategies. InO exploits this mechanism to deliver the DNA-damaging cytotoxin calicheamicin to CD22-positive cells. CD22 is present to a variable extent on B-ALL blasts. The relevance of CD22 protein expression for InO sensitivity has remained controversial in clinical trials, limiting the incorporation of flow cytometry–based CD22 expression thresholds into prognostic models. Escherich et al used a CRISPR screen to identify EBF1 as the most important transcriptional regulator of CD22 expression in B-ALL cells, a finding further confirmed by EBF1 chromatin immunoprecipitation, gene knockouts, and disruption of EBF1 binding sites at the CD22 locus (see figure). The transcription factor EBF1 is essential for B-cell development.6 First expressed in common lymphoid progenitors—which lack typical B-lineage markers—EBF1 initiates B-lineage commitment through coordinated feedback loops with other B-cell transcription factors such as PAX5 and TCF3.6 Remarkably, Escherich et al found that CD22 expression was more dependent on EBF1 than on PAX5, underscoring the specificity and functional significance of the EBF1–CD22 regulatory axis.
The B-cell developmental stage of arrest impacts InO sensitivity in B-ALL samples. Escherich et al used bulk RNA sequencing and a gene expression reference for normal B lymphopoiesis to infer the relative contribution of transformed B-cell developmental stages to individual B-ALL samples. The relative proportion of more immature Pre-pro B cells increased in cases with Ino resistance, while the relative proportion of more mature pro B cells decreases. The InO target antigen CD22 is a direct target for transcriptional regulation by EBF1, which are both initiated at the Pre-pro B stage of normal B lymphopoiesis (CD22 and EBF1 expression taken from https://scminer.stjude.org).7 Professional illustration by Patrick Lane, ScEYEnce Studios.
The B-cell developmental stage of arrest impacts InO sensitivity in B-ALL samples. Escherich et al used bulk RNA sequencing and a gene expression reference for normal B lymphopoiesis to infer the relative contribution of transformed B-cell developmental stages to individual B-ALL samples. The relative proportion of more immature Pre-pro B cells increased in cases with Ino resistance, while the relative proportion of more mature pro B cells decreases. The InO target antigen CD22 is a direct target for transcriptional regulation by EBF1, which are both initiated at the Pre-pro B stage of normal B lymphopoiesis (CD22 and EBF1 expression taken from https://scminer.stjude.org).7 Professional illustration by Patrick Lane, ScEYEnce Studios.
Several groups have come up with gene expression–based definitions of human B lymphopoiesis7-9 as the framework for defining the developmental origins of B-ALL. In taking different methodological approaches, these studies largely concur on the ranking of B-ALL subtypes according to their degree of immaturity, supporting the validity of gene expression–based modeling in this context. Leveraging a developmental gene set definition established by their own group,7 Escherich et al have identified an inferior InO response in a more immature “pre-pro-B-like“ B-ALL cluster, reminiscent of normal Pre-pro-B cells, which also exhibit lower levels of CD22. The CD22 expression itself was not specifically considered in this model, although extremely low EBF1 expression was also associated with decreased CD22 and subsequent InO resistance in this ex vivo treatment cohort.
Molecular B-ALL subtypes also impacted InO sensitivity with more immature subtypes—such as KMT2A-rearranged ALL—showing poorer responses10 in both preclinical and clinical settings. Here, Escherich et al focused on intra-subtype heterogeneity of BCR::ABL1-positive ALL. Using gene expression definitions of multilineage vs lymphoid-only BCR::ABL1 involvement, the authors identified comparatively lower EBF1 and CD22 expression in BCR::ABL1 multilineage cases. This aligns with previous findings indicating a more immature developmental origin of BCR::ABL1 multilineage ALL and corresponds with reduced ex vivo sensitivity to InO in this study.
The efficacy of antibody–drug conjugates depends not only on target gene expression but also on delivery and efficacy of the cytotoxic payload. In this regard, Escherich et al have already shown10 that InO efficacy also depends on expression of the DNA nucleotidylexotransferase (DNTT) and that InO therapy selects for DNTT-low blasts. DNTT plays an important role in the antigen receptor diversification during V(D)J recombination and is regulated by B-cell–specific transcriptional programs, yet it is not a direct transcriptional target of EBF1. Nonetheless, it again seems possible that the B-cell developmental origins of B-ALL samples may contribute to variation in DNTT expression.
One of the limitations of the study is its exclusively preclinical exploration of InO sensitivity. Two B-ALL PDX models, representing the extremes of InO sensitivity, are also used to confirm the range of response phenotypes in vivo. It remains to be seen how B-cell developmental origins shape InO treatment response and clinical outcomes in patients with B-ALL. Will proximity to certain B-cell developmental stages together with EBF1 and CD22 expression be more informative than CD22 protein expression alone? The study does contribute validated gene set definitions and transcription factor targets that could guide the development of predictive models for cell-intrinsic InO resistance, complementing previously described genomically acquired5 modes of resistance. It also raises the broader question of whether the efficacy of other B-ALL immunotherapies, such as Blina or chimeric antigen receptor T cells, may similarly depend on the underlying stage of B-cell developmental arrest. Insights gained from such an analysis would contribute to an integrative risk stratification that could extend beyond the risk factors established for conventional therapy regimens by including also newly established B-cell developmental underpinnings of immunotherapy response.
Conflict-of-interest disclosure: L.B. declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal