In this issue of Blood, Silbert et al1 describe the clinicopathological features and treatment outcomes of 75 cases of lineage switch (LS) in acute leukemia after antigen-targeting immunotherapy. The largest such case series to date, this study highlights the dismal outcomes for most LS patients, reinforcing the need for novel therapeutic approaches, which will need to be informed by a deeper understanding of LS biology. This study provides much needed clinical data to guide clinicians, but it may also establish a framework to guide future prospective collaborative efforts, focused on understanding the mechanisms that drive LS.
B-lymphoid to myeloid LS is characterized by widespread transcriptional and epigenetic reprogramming, leading to inevitable loss of the B-lymphoid antigens CD19 and CD22.2 Initially described as a rare event after chemotherapy,3 LS has emerged as an important resistance mechanism after immunotherapies such as blinatumomab and chimeric antigen receptor T-cell (CAR-T) therapy, which target these antigens. This is particularly the case in B-cell acute lymphoblastic leukemia (B-ALL) with KMT2A rearrangement (KMT2Ar), in which LS after CAR-T therapy occurs with an incidence of up to 24%4 and underscores the inferior post–CAR-T survival observed in this B-ALL subtype.4,5 Prior to this publication, postimmunotherapy LS had been described only in isolated case reports and small case series, and a comprehensive description of its clinical and diagnostic features had been lacking.
This retrospective case series was the product of a large international collaborative effort (Project EVOLVE) spanning 43 sites across 8 countries. The main focus of the study is 70 cases of B-ALL to acute myeloid leukemia (AML) or acute leukemia of ambiguous lineage (ALAL) LS. Classification as LS required a change in World Health Organization, International Consensus Classification, and/or European Group of Immunological Leukemia classification from B-ALL to AML or ALAL, along with evidence of clonal relatedness provided by the presence of concordant genomic characteristics, or B-cell receptor (BCR) sequences. Although the authors used a uniform set of criteria to define LS, immunophenotyping methods applied at diagnosis and relapse were variable across sites, and it may not have been possible to exclude an initial diagnosis of ALAL in some patients.
Most LS occurred after CAR-T therapy (49%) or blinatumomab (44%), along with 4 instances after inotuzumab ozogamicin and a sole case after treatment with an investigational CD19 antibody drug conjugate (ADC). Unsurprisingly, the majority of the cohort comprised KMT2Ar B-ALL (64%). The remainder of cases included non-KMT2Ar subtypes that have appeared previously in the LS literature (eg, ZNF384r, BCR::ABL1); however, multiple previously unreported B-ALL subtypes were also identified, including CRLF2r, MEF2Dr, IGH::IL3, and TCF3::HLF B-ALL, and 3 patients with B-ALL, not otherwise specified, which harbored TP53 mutations. Outside of subtype-defining driver lesions, molecular characterization was not sufficiently uniform to draw conclusions surrounding the potential role cooperating genomic events play in LS pathogenesis.
Post-LS outcomes were poor. Median overall survival after LS was just 4.8 months and <10% of patients experienced long-term disease-free survival. Treatment strategies were heterogeneous, and no approach led to frequent responses. Intensive chemotherapy demonstrated a complete response (CR) rate of just 26%, and responses were rare among the few patients who received novel therapies. The low CR rate to venetoclax-azacitidine (33%, 2 of 6) may reflect the high frequency in this cohort of post-LS monocytic phenotypes, which has been associated with venetoclax resistance.6 Disappointingly, menin inhibitors, which have shown promising activity in de novo AML with KMT2Ar,7 yielded no responses (CR 0%, 0 of 6), and it would be of interest to consider how the biology of post-LS KMT2Ar AML may differ from its de novo counterpart. The anti-CD33 ADC gemtuzumab ozogamicin, administered in combination with chemotherapy, was associated with the highest CR rate (50%, 7 of 14); however, the small sample size and heterogeneous nature of the cohort preclude meaningful comparisons or firm treatment recommendations.
Given the difficulty in salvaging LS once it has occurred, potential preemptive strategies to prevent LS after immunotherapy may be a more attractive approach. Consolidative allogeneic stem cell transplant (alloSCT) was a component of therapy in 4 of the 5 long-term survivors and may have efficacy against LS-prone clones. However, given the rapid development of LS (median 1.5 months, >80% within 6 months), this may not be feasible as a preemptive, postimmunotherapy strategy for the majority of patients in whom LS is destined to occur.
To identify those at highest risk, and design effective therapies to treat or prevent postimmunotherapy LS, it will be essential to better understand its biology (see figure). Two main origins of LS have been proposed2,8: (1) myeloid relapse arising from a primitive precursor-like population without interval lymphoid differentiation; and (2) transdifferentiation of previously committed B-ALL cell directly to AML. Although not the intent of this study, BCR sequencing (predominantly using the clonoSEQ platform) performed for diagnostic purposes in 25 patients is hypothesis-provoking. Of these patients, 80% demonstrated persistence of the same clonotype between the initial B-ALL and subsequent AML/ALAL, suggesting that transdifferentiation is a major contributor to LS after immunotherapy.
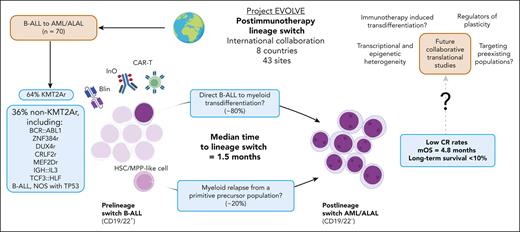
Postimmunotherapy lineage switch. In the international Project EVOLVE cohort, postimmunotherapy lineage switch (LS) occurred primarily in KMT2A-rearranged B-ALL; however, multiple other B-ALL subtypes were represented. LS occurred after CD19 and/or CD22 targeting CAR-T therapy, blinatumomab and inotuzumab ozogamicin---it remains unclear whether immunotherapy directly induces LS or selects for preexisting myeloid or less differentiated clones. B-cell receptor sequencing of pre- and post-LS leukemia suggests that LS may occur via direct B-ALL to myeloid transdifferentiation, but it has also been proposed to arise from primitive precursor populations. LS occurs rapidly, and post-LS treatment outcomes are dismal. Structured collaborative translational research to address the mechanisms that drive LS will likely lead to improved outcomes. ALAL, acute leukemia of ambiguous lineage; AML, acute myeloid leukemia; B-ALL, B-cell acute lymphoblastic leukemia; Blin, blinatumomab; CAR-T, chimeric antigen receptor T cell; HSC, hematopoietic stem cell; Ino, inotuzumab ozogamicin; LS, lineage switch; NOS, not otherwise specified; mOS, median overall survival; MPP, multipotent progenitor cell.
Postimmunotherapy lineage switch. In the international Project EVOLVE cohort, postimmunotherapy lineage switch (LS) occurred primarily in KMT2A-rearranged B-ALL; however, multiple other B-ALL subtypes were represented. LS occurred after CD19 and/or CD22 targeting CAR-T therapy, blinatumomab and inotuzumab ozogamicin---it remains unclear whether immunotherapy directly induces LS or selects for preexisting myeloid or less differentiated clones. B-cell receptor sequencing of pre- and post-LS leukemia suggests that LS may occur via direct B-ALL to myeloid transdifferentiation, but it has also been proposed to arise from primitive precursor populations. LS occurs rapidly, and post-LS treatment outcomes are dismal. Structured collaborative translational research to address the mechanisms that drive LS will likely lead to improved outcomes. ALAL, acute leukemia of ambiguous lineage; AML, acute myeloid leukemia; B-ALL, B-cell acute lymphoblastic leukemia; Blin, blinatumomab; CAR-T, chimeric antigen receptor T cell; HSC, hematopoietic stem cell; Ino, inotuzumab ozogamicin; LS, lineage switch; NOS, not otherwise specified; mOS, median overall survival; MPP, multipotent progenitor cell.
In the study, LS occurred after an MRD-negative response in >50% of patients, suggesting that it often arises from a rare leukemia subpopulation. Studies using single-cell techniques in KMT2Ar B-ALL have identified candidate populations of B-lymphoid cells that appear epigenetically primed for myeloid differentiation.9 Such a population may serve as a reservoir of transdifferentiation potential; however, it remains unclear how immunotherapy itself may interact with tumor heterogeneity and plasticity. Does therapy simply select for preexisting primitive or myeloid clones lacking CD19 and/or CD22 expression? Or may it provide specific extrinsic differentiation signals that directly promote LS in primed leukemic subpopulations?10
Understanding the cellular origins of LS, the regulators of intratumor heterogeneity and plasticity, and how immunotherapy-derived signals interact with these tumor-intrinsic factors is a necessary step toward developing effective therapies to prevent and treat LS. Such discoveries can only be achieved through concerted collaboration between clinical, translational, and basic science researchers, and must be supported by rigorous, prospective biobanking along with uniform and considered experimental approaches. B-ALL has a long and fruitful history of academic collaboration, and it is hoped that the rich clinical data and international collaborative framework established by the Project EVOLVE investigators can be leveraged to illuminate and eventually overcome this vexing phenomenon.
Conflict-of-interest disclosure: J.A.K. declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal