In this issue of Blood, Andricovich et al1 demonstrated that BAP1 and TP53 deficiencies cooperate to promote the transformation of erythroid-primed multipotent progenitors toward erythroleukemia or mixed acute myeloid leukemia (AML). They further revealed a dependency on BCL-xL (B-cell lymphoma extra large) in BAP1/TP53–deficient erythroleukemia, thereby uncovering a potential therapeutic vulnerability.
Although TP53 mutations are found in fewer than 10% of de novo AML cases, these patients have a poor prognosis with a 1-year overall survival rate of less than 20%.2-4TP53 mutations rarely co-occur with other known myeloid transformation drivers, yet they are frequently linked to complex karyotypes.2,4,5 Although biallelic TP53 mutations define pure erythroid leukemia (PEL), acute erythroid leukemia (AEL), which contains fewer nucleated erythroid cells, may also harbor TP53 mutations and share a similar poor prognosis.3,6 However, the cooperative genetic events that shape erythroid progenitors in TP53-mutated AML and the associated therapeutic vulnerabilities remain incompletely understood. Andricovich et al identified that BAP1 loss is a cooperative event with TP53 deficiency that transforms erythroid-primed multipotent progenitors and revealed that there is a BCL-xL dependency in BAP1/TP53–deficient erythroleukemia (see figure).
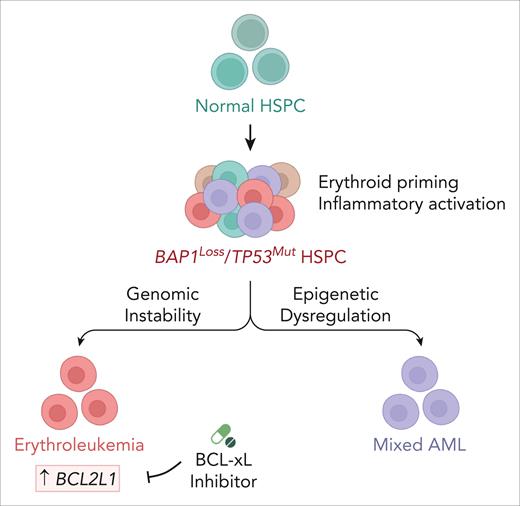
The loss of BAP1 in TP53-mutated AML drives the transformation of erythroid-primed HSPCs and is accompanied by transcriptional reprogramming and inflammatory activation. Genomic instability and epigenetic dysregulation contribute to lineage divergence toward erythroleukemia or mixed AML, respectively. BCL-xL inhibition selectively targets BAP1/TP53–deficient erythroleukemia, thereby reducing the leukemic burden and partially restoring erythropoiesis. The schematics were created with BioRender.com.
The loss of BAP1 in TP53-mutated AML drives the transformation of erythroid-primed HSPCs and is accompanied by transcriptional reprogramming and inflammatory activation. Genomic instability and epigenetic dysregulation contribute to lineage divergence toward erythroleukemia or mixed AML, respectively. BCL-xL inhibition selectively targets BAP1/TP53–deficient erythroleukemia, thereby reducing the leukemic burden and partially restoring erythropoiesis. The schematics were created with BioRender.com.
Although previous studies have linked the progression of TP53-mutated preleukemic clones to AML with erythroid immunophenotypes and inflammatory features,7 the molecular mechanisms that underlie erythroleukemia transformation have not been elucidated fully. The study showed that BAP1 loss occurs in approximately one-third of TP53-mutated AML cases and is associated with an erythroid-primed transcriptomic profile and even poorer overall survival than those without a BAP1 loss. BAP1 encodes a deubiquitinase that is involved in epigenetic regulation through the removal of H2AK119Ub and forms the Polycomb repressive deubiquitinase complex with ASXL1 (Additional sex combs–like 1).8,9 Using transgenic mouse models, the authors showed that the combined loss of BAP1 and Trp53 (Bap1;Trp53DKO) drives the development of erythroleukemia from heterogeneous hematopoietic stem and progenitor cell (HSPC) populations, which leads to either PEL, AEL, or, in some instances, mixed AML. Inducible depletion of BAP1 and Trp53 in adult mice (Mx1Cre;Bap1 fl/fl;Trp53 fl/fl) similarly leads to erythroleukemia, and the consequently transformed cells demonstrate robust leukemic reconstitution and competitive advantage upon transplantation, closely resembling human disease.
One of the notable strengths of this study lies in its comprehensive dissection of the transcriptional and chromatin dynamics across leukemia subtypes by using an array of bulk and single-cell approaches. Intriguingly, the authors found that PEL retains a mature erythroid transcriptional signature, whereas AEL preserves features of erythroid-primed multipotent progenitors, underscoring the transcriptional heterogeneity of Bap1;Trp53DKO leukemias. To further elucidate the molecular basis of this lineage divergence, the authors conducted single-cell transcriptomic and epigenomic profiling. Either BAP1 or Trp53 deficiency can activate proinflammatory programs in HSPCs and multipotent progenitors (MPPs), whereas their combined loss leads to a pronounced erythroid differentiation block and aberrant expansion of erythroid progenitors. Epigenetic dysregulation diverts erythroid-primed progenitors toward a myelomonocytic fate, thereby contributing to the development of mixed AML. In contrast, genomic instability lowers the threshold for malignant transformation along the erythroid lineage, thereby promoting erythroleukemia.
Beyond mechanistic insights, the study uncovered a selective dependency on BCL-xL in BAP1/TP53-deficient erythroleukemia, reinforcing the translational relevance of the findings. The authors identified that BCL2L1 is upregulated in BAP1-deficient AML patients and in murine Bap1;Trp53DKO erythroleukemia. BCL-xL inhibition with A-1155463 mitigated the leukemic burden and selectively reduced the erythroid-primed MPP2 and proerythroblast populations in Bap1;Trp53DKO mice, suggesting partial recovery of erythropoiesis. Single-cell RNA sequencing analysis further revealed that A-1155463 treatment in Bap1;Trp53DKO PEL mice restored the frequency of HSPCs and erythroid progenitors, induced myelomonocytic expansion, and attenuated proinflammatory signaling. In addition, it reduced the number of genomically unstable subclones and lowered the transcriptional complexity in neoplastic erythroid progenitors. Despite the inherent challenges of studying TP53-mutated erythroleukemia, Andricovich et al have made a significant contribution by elucidating its pathogenic mechanisms and uncovering a therapeutic vulnerability that may benefit genetically defined AML subtypes. They demonstrated that BAP1 loss cooperates with TP53 deficiency to transform erythroid-primed multipotent progenitors, which leads to distinct leukemic phenotypes, and identified that BCL-xL is a promising therapeutic target, thereby opening new avenues for understanding disease mechanisms and clinical intervention. Although these findings laid pivotal groundwork, several challenges remain. It will be important to validate the efficacy of BCL-xL inhibition in human AML samples, assess its long-term effects, and determine how it might be effectively integrated with the current therapeutic regimens. Future research that addresses disease heterogeneity and potential mechanisms of treatment resistance will be critical to advancing this approach toward clinical translation.
Conflict-of-interest disclosure: The authors declare no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal