In this issue of Blood, Singh et al report a 2.4-Å resolution cryogenic electron microscopy (cryo-EM) structure of coagulation factor XIII (FXIII).1 The structure finally reveals the architecture of the A2B2 heterotetrameric assembly, advances basic knowledge, and offers molecular context for the clinical manifestations of FXIII deficiency.
Coagulation FXIII is a transglutaminase essential to the response to vascular injury. Once activated by thrombin in the last step of the coagulation cascade, it cross-links fibrin polymers and produces a hemostatic plug that traps red blood cells and platelets, thereby determining thrombus stability, composition, and size.2 The proenzyme form of FXIII is composed of 2 tightly bound homodimers of independently synthesized subunits, A and B, responsible for catalytic activity (A subunit) and regulation (B subunit). The resulting A2B2 heterotetramer circulates in the blood bound to fibrinogen,3 an interaction that promotes binding of thrombin and conversion of FXIII to its active form through cleavage of the A subunit in a Ca2+-dependent manner. Available structures of the A subunit have detailed both the zymogen and active forms and identified the Ca2+-binding sites responsible for activation.4,5 The A subunit is composed of a β-sandwich domain, a core domain containing the catalytic C315, and 2 terminal β-barrel domains. The B subunit consists of 10 Sushi domains homologous to those of complement factors and features considerable conformational flexibility, as documented in previous biophysical studies using low-resolution EM6 and atomic force microscopy.7 No structure of the B subunit is currently available, but its molecular cross talk with the A subunit is well established and of considerable functional significance.8
Singh et al advance basic knowledge of FXIII by describing a long-awaited structure of the A2B2 heterotetramer as a significant segue to recent modeling studies by the same group.9 The structure, solved by cryo-EM at 2.4-Å resolution, reveals the heterotetramer with a completely resolved A2 dimer in complex with Sushi domains 1 and 2 of the B2 subunits. A second, 3.0-Å resolution structure documents additional information on Sushi domains 3 through 5. Sushi domains 6 through 10 remain unresolved, consistent with their dynamic nature, as reported elsewhere.6,7
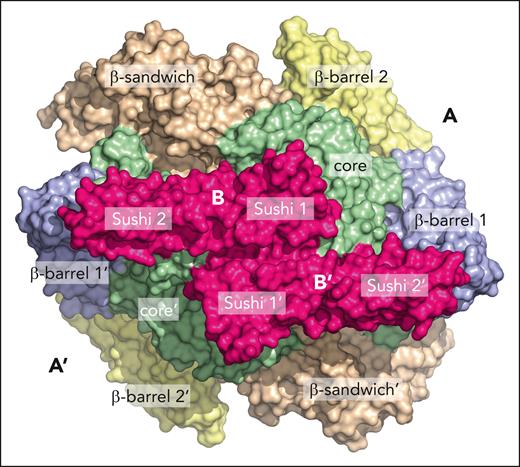
The A2B2 heterotetramer assumes a crownlike shape, with the A2 dimer at the bottom supporting the B2 dimer at the top with 2 arms joining in the center (see figure). The structure is largely symmetric, except for the interactions among Sushi domains 1, 4, and 5 across the B-B interface that introduce a degree of asymmetry and conformational disorder that could easily extend to Sushi domains 6 through 10 not visible in the density map. Within the heterotetramer, the A2 subunits show no significant structural difference from previously reported crystal structures,4,5 indicating a rigid-body association with the B2 dimer. The A2 dimer has a convex base bearing the N-terminal activation peptide cleaved by thrombin and a bowl-shaped face binding the B2 dimer. This arrangement masks the Ca2+-binding sites II and III linked to activation and leaves the regulatory Ca2+-binding site I free. The catalytic triad (C315, H374, D397) resides on the convex base of the A2 dimer within the A-A interface and away from the B-B interface. In the B subunits, the resolved Sushi domains exhibit the typical 3+2 β-strand arrangement seen in many Sushi domain–containing proteins.
Interfaces A-A, B-B, and A2-B2 deserve attention. The A-A interface contains 25 salt bridges and 33 H bonds, mostly contributed by the catalytic core. Each B subunit interacts with the other B subunit through several Sushi domains and with both A subunits through Sushi domains 1 and 2 in an antiparallel configuration (see figure). Notably, the catalytic core of the A domain interacts with Sushi domains 1 and 2 mainly through hydrophobic or polar contacts, respectively, but the A2-B2 interface is stabilized by electrostatic interactions. The A2B2 architecture explains how binding of the B subunits to the concave side of the A2 dimer buries the secondary thrombin cleavage site at K514 and enhances the half-life of the A2B2 heterotetramer. On the other hand, plasmin cleavage at K468 and Q469 would require disruption of both the A2-B2 and A-A interfaces after activation, accounting for the short half-life of the activated A subunit in the blood.
Cryo-EM structure of the A2B2 heterotetramer of FXIII showing the arrangement of the 2 A subunits, with the 4 constitutive domains and the 2 B subunits containing Sushi domains 1 and 2 visible in the high-resolution (2.4-Å) structure. Note the highly symmetric assembly with the B subunits bridging the core domains of the A subunits.
Cryo-EM structure of the A2B2 heterotetramer of FXIII showing the arrangement of the 2 A subunits, with the 4 constitutive domains and the 2 B subunits containing Sushi domains 1 and 2 visible in the high-resolution (2.4-Å) structure. Note the highly symmetric assembly with the B subunits bridging the core domains of the A subunits.
Mapping known missense mutations on the structure of FXIII offers insight into the molecular origin of observed bleeding phenotypes. New in-frame deletions and missense mutations are also reported in the study. Interestingly, the homozygous missense mutations tend to distribute across the complex, whereas the heterozygous missense mutations cluster around the A-A, B-B, and A2-B2 interfaces, with only Q416R occurring at the A-B heterodimeric interface. Most missense mutations affect the A subunit in the catalytic core, away from the A2-B2 interface, but R409L is near the A-A interface and Ca2+-binding site III. A crude, in silico assessment of the disruptive effect of these mutations shows that 3 of 17 reported heterozygous missense variants are disruptive, compared with 5 of 65 homozygous ones. The difference may explain the dominant negative etiology of heterozygous missense variants in mild FXIII deficiency.
In addition to offering an interpretation of molecular defects associated with bleeding phenotypes and providing a platform for the design of new therapeutic strategies, the cryo-EM structure of FXIII validates important aspects of FXIII biochemistry and opens new opportunities for the field. An unsolved issue is how the A2B2 heterotetramer engages fibrinogen, whose epitope recognizing FXIII has been mapped to the short 390-396 segment of the γ chain.3,10 The reciprocal surface of interaction on FXIII is not known but presumably involves 1 or more of the Sushi domains.10 A complex of FXIII with fragments of fibrinogen comprising the 390-396 segment of the γ chain should be considered for cryo-EM analysis. Finally, Singh et al provide the latest testimony to the power of cryo-EM in solving structures that have eluded X-ray crystallographers for decades. Investment in this technique would be a wise move for those who wish to advance our understanding of coagulation factors at the molecular level.
Conflict-of-interest disclosure: E.D.C. declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal