In this issue of Blood, Chapuy et al report on the development of DLBclass,1 a probabilistic, neural network–based classifier that assigns individual cases of diffuse large B-cell lymphoma (DLBCL) into 1 of 5 genetic subtypes, as described previously by the same group.2
DLBCL is a molecularly heterogeneous disease. This heterogeneity limits our ability to implement successful precision medicine strategies in DLBCL treatment.3 Two main transcriptional subtypes of DLBCL were first described more than 2 decades ago, but lacked the granularity required to guide biologically targeted therapies.4 More recently, genetic subtypes of DLBCL have emerged from exome and targeted sequencing studies. Those studies each clustered large cohorts of DLBCL into 5 to 7 genetic subtypes, with 3 independent groups converging onto remarkably similar classification systems.2,5,6 Early data suggest that these genetic subtypes may predict response to biologically targeted therapies.7,8 A requirement to exploit these classification systems, derived from clustering of large cohorts, is a classifier tool to assign subtypes to an individual biopsy. This was first addressed by LymphGen, a probabilistic classifier that assigns 1 of 7 genetic subtypes to an individual case of DLBCL.9 A limitation of the LymphGen is that approximately 40% of cases remain in an unclassified or composite category. To address this, Chapuy et al describe the development of a new classifier tool based on their own classification system wherein every patient with detectable mutations is assigned a genetic subtype.
The authors first validated their previous classification system using an expanded cohort of 699 patients that included cases from the Schmitz et al study.5 The cohort was divided into 550 cases used to train a neural network–based probabilistic classifier, and 149 cases were used as an independent validation cohort to test classifier performance. The final classifier, which takes input from single-nucleotide variants, somatic copy number alteration, and structural variants, assigned all cases in the validation cohort to a genetic subtype with 89% accuracy. Greater accuracy (98%) was seen in the 74% of cases in which the classification confidence was >0.7. Performance was degraded if the full genetic data were not available, such as if the input data were restricted to that provided by other commonly used sequencing panels, or if either somatic copy number alteration or structural variant data were unavailable. When compared with LymphGen, there was a high level of classification to the analogous genetic subtype. As expected, agreement was highest amongst the cases assigned with high confidence (>0.7) by DLBclass. All cases previously unclassified by LymphGen were assigned a genetic subtype by DLBclass. Importantly, the authors have made DLBclass freely available as an easy-to-use online tool.
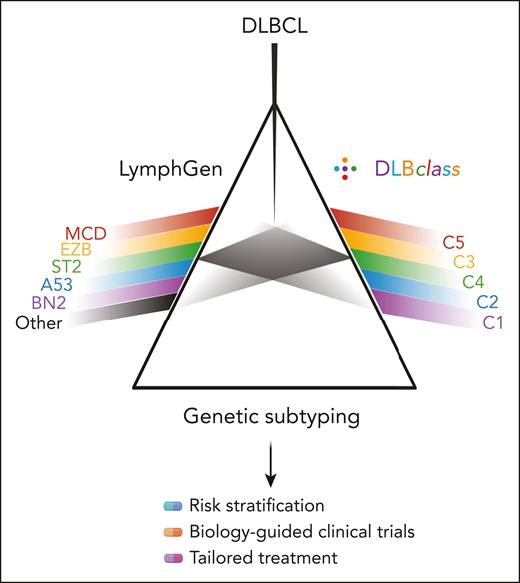
There are now 2 classifiers available to DLBCL researchers and clinicians (see figure). Superficially, these classifiers give similar answers, but there are important differences. The LymphGen NOTCH1-mutated (N1) or double-hit–like (EZB-MYC) subtypes are not recognized by DLBclass. However, these cases are relatively rare and easily identified by other methods. The input data are different. DLBclass requires single-nucleotide variant data to be entered as a binary (mutant or not mutant) gene level call, and this was trained and tested on data run through the authors’ own harmonized variant calling and annotation pipeline. Alternative pipelines could lead to a slightly different genotyping output that may affect classifier performance. However, the most striking difference is how each classifier handles those cases that do not fit robustly into one of the prototypical subtypes. This leads to a striking difference in the proportion of cases that can be classified; that is, up to approximately 60% with LymphGen, but 100% for DLBclass. DLBclass takes the view that every case of DLBCL can be classified into 1 of 5 genetic subtypes. It assigns the most likely case regardless of confidence level. This is certainly very attractive to those who plan to run precision medicine trials of subtype-directed therapies. Indeed, such a trial would need to recruit almost twice as many patients if stratified by LymphGen. In contrast, LymphGen takes the view that some cases of DLBCL must belong in presently undiscovered subtypes, or simply cannot be classified with sufficient confidence from the available genetic data. Over-assigning these cases may dilute the molecular purity of individual subtypes, stymying the potential to resolve subtype-specific responses to targeted therapy in a precision medicine trial. At present, there is no good way to determine which of these approaches to DLBCL classification is optimal. There is no ground truth as to the molecular subtype of an individual case. The answer will instead come from the application of these approaches to clinical trial cohorts, with the optimal classification being the one that best predicts response to targeted therapies. When applied retrospectively to the Phoenix trial, albeit with genetic information available for very small patient numbers, LymphGen distinguished subtypes that benefited from the addition of ibrutinib to R-CHOP therapy.7 Unfortunately, the available targeted sequencing data contain too few of the genetic features needed to apply DLBclass. This leaves us in the extremely disappointing position that, despite many thousands of patients having been enrolled in randomized trials of targeted therapies in DLBCL, there is not a single trial in which the molecular data required to run both LymphGen and DLBclass were both collected and made available. Those designing, funding, or publishing prospective clinical trials of DLBCL should strive to ensure they collect, and then make available, the appropriate molecular data required to apply current classifiers. Researchers should also be aware that DLBCL classification is a moving target, evolving on multiple fronts that include transcriptional, microenvironmental, and proteomic subtypes of DLBCL. This makes it essential that acquired molecular data be broad enough to apply future emergent classification systems. Without this anticipation, a classification system may take years or more to have its value established.
The discovery of genetic subtypes of DLBCL is a major advance in taming the molecular heterogeneity of DLBCL, which otherwise acts as a barrier to precision medicine. Having 2 classifiers that take slightly different approaches to genetic classification of DLBCL is exciting and takes us one step closer to the promise of precision medicine in DLBCL treatment. Their value will ultimately be judged by their ability to predict responses to targeted therapies in future clinical trials.
Parallel genetic classifiers now exist for DLBCL. DLBclass joins LymphGen as a classifier that assigns individual cases of DLBCL to genetic subtypes. Corresponding subtypes across the 2 classifications are shown in matching colors. The most important distinction relates to the handling of cases that cannot be classified with high confidence. The relative predictive value of each classifier will emerge from their application in future clinical trials. Image created in BioRender.com.
Parallel genetic classifiers now exist for DLBCL. DLBclass joins LymphGen as a classifier that assigns individual cases of DLBCL to genetic subtypes. Corresponding subtypes across the 2 classifications are shown in matching colors. The most important distinction relates to the handling of cases that cannot be classified with high confidence. The relative predictive value of each classifier will emerge from their application in future clinical trials. Image created in BioRender.com.
Conflict-of-interest disclosure: D.J.H. received research funding from AstraZeneca and GlaxoSmithKline. J.A.K. declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal