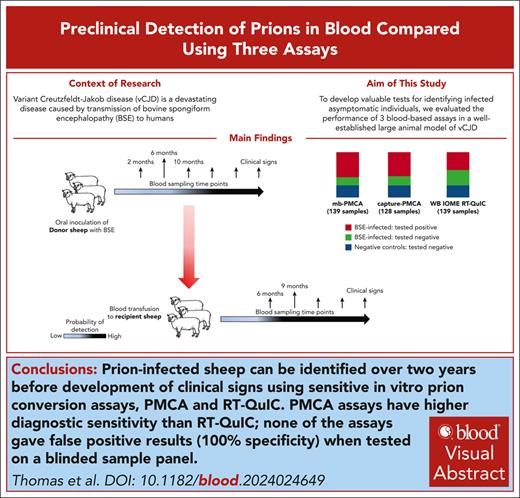
Key Points
Three blood tests detected prion infection in sheep blood samples up to 26 months before the onset of clinical signs, with 100% specificity.
The tests are applicable to human variant Creutzfeldt-Jakob brain samples and may be adaptable for prion detection in human blood.
Visual Abstract
Variant Creutzfeldt-Jakob disease (vCJD) is a devastating disease caused by transmission of bovine spongiform encephalopathy to humans. Although vCJD cases are now rare, evidence from appendix surveys suggests that a small proportion of the United Kingdom population may be infected without showing signs of disease. These “silent” carriers could present a risk of iatrogenic vCJD transmission through medical procedures or blood/organ donation, and currently there are no validated tests to identify infected asymptomatic individuals using easily accessible samples. To address this issue, we evaluated the performance of 3 blood-based assays in a blinded study, using longitudinal sample series from a well-established large animal model of vCJD. The assays rely on amplification of misfolded prion protein (PrPSc; a marker of prion infection) and include real-time quaking-induced conversion (RT-QuIC), and 2 versions of protein misfolding cyclic amplification (PMCA). Although diagnostic sensitivity was higher for both PMCA assays (100%) than RT-QuIC (61%), all 3 assays detected prion infection in blood samples collected 26 months before the onset of clinical signs and gave no false-positive results. Parallel estimation of blood prion infectivity titers in a sensitive transgenic mouse line showed positive correlation of infectivity with PrPSc detection by the assays, suggesting that they are suitable for detection of asymptomatic vCJD infection in the human population. This study represents, to our knowledge, the largest comparison to date of preclinical prion detection in blood samples from a relevant animal model. The outcomes will guide efforts to improve early detection of prion disease and reduce infection risks in humans.
Introduction
Prion diseases are fatal neurodegenerative disorders of humans and animals,1,2 featuring templated conversion of a cellular glycoprotein, “prion protein” (PrPC), into a misfolded isoform (PrPSc)3 as a key pathogenic process. PrPSc accumulates in the brain and other tissues,4-6 and is believed to represent the infectious agent responsible for disease transmission.3 The most common human prion disease is sporadic Creutzfeldt-Jakob disease (CJD),7 which appears not to have an infectious etiology. Variant CJD (vCJD) emerged after human exposure to products from cattle infected with bovine spongiform encephalopathy (BSE),8 and remains the only known example of zoonotic transmission of prions.9-12
Infectious prion diseases have long asymptomatic incubation periods, which can range from several months to years. During this period, infected individuals are at risk of transmitting infection to others; indeed, a number of cases of vCJD are attributed to transfusions with blood products from asymptomatic infected donors.13-16 Diagnosis of prion disease can generally only be confirmed post mortem, usually by detection of PrPSc in brain sections or extracts. Despite strenuous research efforts, a high-throughput assay capable of reliably detecting preclinical prion infection using readily accessible samples (such as blood or urine) has proven elusive. This is partly because the concentration of PrPSc in body fluids is very low17,18; accordingly, recent technical developments have used amplification and/or selective concentration of PrPSc to overcome this issue.19-27
Cell-free prion amplification assays, such as protein misfolding cyclic amplification (PMCA) and real-time quaking-induced conversion (RT-QuIC), exploit the ability of PrPSc to self-propagate through template-directed misfolding.28-30 In 2 independent blinded studies,24,31 PMCA assays identified blood samples from patients with vCJD with 100% sensitivity and specificity.24 RT-QuIC is now routinely used on cerebrospinal fluid samples as a diagnostic test for sporadic CJD.32 Although RT-QuIC has also been shown to detect PrPSc in blood and body fluids of animals naturally and experimentally infected with prions,19-21,23,26,27,33,34 there is no published data on its application to blood samples from patients with vCJD. We recently showed that with inclusion of an iron oxide magnetic extraction (IOME) step,23 RT-QuIC could detect PrPSc in whole blood (WB) from BSE-infected sheep, including samples from the preclinical stage of infection.35
Blood-based diagnostic tests would be of greatest benefit if they were able to detect preclinical prion infection, enabling prevention of transmission and delivery of treatments when they are likely to be most effective. Because there are minimal numbers of blood samples available from presymptomatic vCJD cases, preclinical samples from animal models offer an alternative for initial evaluation of test performance. Our previous work established BSE-infected sheep as a model to study the transmission of vCJD by blood transfusion,36 and produced an extensive archive of blood samples from individual infected sheep. Here, we describe a comparison of the detection of PrPSc by 3 in vitro assays in longitudinal series of blood samples from infected sheep, using blinded panels of samples from the archive. The assays evaluated included miniaturized bead PMCA (mb-PMCA),36-38 plasminogen bead capture PMCA (capture-PMCA),24,39 and WB IOME RT-QuIC.35 Assay results were correlated with blood infectivity titers estimated by intracerebral inoculation of transgenic mice overexpressing bovine PrP (TgBov mice).
Methods
Reference brain homogenates
The source and preparation of reference brain homogenates from BSE-infected sheep (BSB/7/10) and a variant CJD case (reference NHBY0/0003; codon 129 MM) have previously been described in detail.35
Source of sheep blood samples
Blood samples were sourced from an archive established during blood transfusion experiments performed using BSE-infected sheep, which have previously been described in detail.36,40,41 The archive, which was split between the National Institute for Biological Standards and Controls, now Medicines and Healthcare Products Regulatory Agency (NIBSC/MHRA), and The Roslin Institute, contained multiple aliquots of WB, plasma, buffy coat, and red cells collected, using EDTA as anticoagulant and stored at −80°C, from individual infected and control sheep at regular intervals throughout infection.
Preparation of blinded panels
Three panels each containing 128 blood samples from the same sheep and time points were assembled from the NIBSC/MHRA archive in a random order in unlabeled tubes. The panels included samples previously analyzed in an unblinded study using mb-PMCA.36 Each panel contained 94 blood samples from known BSE-infected sheep and 34 samples from negative control (mock-infected) sheep, and were shipped on dry ice to participating scientists/laboratories. Buffy coat panels were tested by mb-PMCA and capture-PMCA; whole-blood samples were tested by WB IOME RT-QuIC. The results were submitted to NIBSC/MHRA for unblinding and decoding.
An additional blinded panel, containing 8 samples from BSE-infected sheep and 3 samples from negative control sheep collected at 10 months post inoculation (mpi), was also tested independently by mb-PMCA and WB IOME RT-QuIC. Because these samples were not included in the NIBSC/MHRA archive, they were prepared, unblinded, and decoded by a scientist unconnected to the project at The Roslin Institute. Because of the French moratorium on prion research from July 2021 to 2024, this panel could not be analyzed by capture-PMCA.
Bioassay of sheep blood samples
Mouse experiments were approved by the local École Nationale Vétérinaire de Toulouse ethics committee and performed in compliance with the European Directive 2010/63/EU. The scientists conducting the bioassay experiments were blinded to sample identity until the conclusion of the study. Briefly, groups (n = 12) of 6- to 10-week-old female transgenic mice overexpressing bovine PrP (TgBov-tg110)18,38,42,43 were anesthetized with isoflurane and inoculated intracerebrally with 20 μL of undiluted buffy coat homogenate, as previously described.42 Observation and assessment for clinical signs of prion disease were performed as previously described,42 and animals were euthanized at predefined humane end points. The brain was removed, 1 half was fixed in 10% formal saline, and the other half frozen at −20°C. If clinical signs were not observed, mice were euthanized at the end of their natural lifespan (∼600-750 days), and brains collected as described above. For each animal, brain samples were analyzed for PrPSc detection by western blot using the Sha31 PrP-specific monoclonal antibody, as previously described.38 The infectious titer in a reference 10% (weight to volume) brainstem homogenate from a sheep with clinical BSE was estimated by the Spearman-Kärber method after end point titration (intracerebral route) in TgBov mice. These data were used to estimate titers of prion infectivity in BSE-infected sheep blood samples (infectious dose causing infection in 50% injected animals [ID50]/mL buffy coat) by a previously reported method44 (supplemental Figure 1, available on the Blood website). Titers were converted to ID50/mL WB by multiplying by 0.1, because the volume of buffy coat is ∼10% of the whole-blood volume.41
mb-PMCA
The mb-PMCA protocol was performed as previously described.36-38 The source of PrPC substrate was brains of healthy transgenic TgShpXI mice (overexpressing sheep PrPC with alanine, arginine and glutamine at codons 136, 154, and 171, respectively).45 PMCA reactions, seeded with 5 mL undiluted buffy coat or brain homogenate, were performed in 96-well polymerase chain reaction microtiter plates placed in a microplate horn attached to a programmable Misonix Q700 sonicator (Qsonica).36 Reaction products were analyzed for detection of PrPSc by western blot with the ROS-BC6 monoclonal antibody.46
Each buffy coat sample from the blinded panel was run in triplicate in 2 independent mb-PMCA experiments. Samples were categorized as positive if ≥2 of 3 replicates were positive in both experiments, or “weak positive” if results were discordant between runs (eg, negative in 1 run, and positive in at least 1/3 replicates in the other). Positive controls were serial 10-fold dilutions of the reference BSE-infected sheep brain homogenate (BSB/7/10) and buffy coat from a sheep clinically affected with BSE. Negative control samples consisted of equivalent samples from mock-infected sheep.
capture-PMCA assay
The testing procedure was developed for the diagnosis of vCJD in plasma samples.24 Briefly, buffy coat samples (25 μL) were digested with benzonase (50 U/mL) for 30 minutes at 37°C in 400 μL of ligation buffer; 10 μL of plasminogen-coated magnetic beads (10 μg plasminogen/mg of beads) were added and incubated for 90 minutes at 25°C. After serial washing steps with phosphate-buffered saline, beads were resuspended in PMCA substrate (90 μL) supplemented with heparin (100 μg/mL). PrPC substrate was brain homogenate from healthy transgenic mice overexpressing human PrP with methionine at codon 129 (Tg650 line).47 Amplification (successive rounds of 80 cycles) was performed in a Q700 sonicator (Qsonica), as previously described.24 Reaction products were analyzed for PrPSc by western blot with the 3F4 monoclonal antibody.24
Samples from the blinded panel were run in duplicate, and a positive result recorded if PrPSc was detected in both experimental replicates in 2 independent experiments. Positive controls included in each capture-PMCA assay were 10-fold dilutions of the reference BSE-infected sheep brain homogenate (BSB/7/10), spiked into 25 μL of buffy coat from healthy human blood donors. Each experiment also included unseeded samples and samples seeded with buffy coat from healthy human blood donors as negative controls.
WB IOME RT-QuIC assay
The WB IOME RT-QuIC assay was performed as previously described.35 Aliquots of WB were thawed at room temperature and 200 μL was diluted in capture buffer containing iron oxide beads, before overnight incubation at room temperature with rotation. After washing and sonication, the beads were magnetically separated and resuspended in 20 μL 0.025% sodium dodecyl sulfate–phosphate-buffered saline. Resuspended beads (2 μL) were used to seed RT-QuIC reactions, using previously described reaction conditions.35 The recombinant PrPC substrate (recPrP) was N-terminally truncated ovine recPrP (ARQ allele; residues 94-233). WB IOME RT-QuIC experiments were assigned a cutoff time of 20 hours, and threshold fluorescence was set at 75% of maximum fluorescence reading for the plate. Samples were assayed in replicates of 4 and were considered positive if ≥2 of 4 replicate reactions exceeded the threshold within 20 hours.
Blinded panels of whole-blood samples were tested in 2 independent experiments; samples that gave inconsistent results (n = 27) were tested a third time, with the outcome determining the final status (positive or negative). Negative controls included unseeded reactions, reactions seeded with a 10−4 dilution of uninfected sheep brain homogenate, and reactions seeded with uninfected blood. Plates were rejected if >1 of 10 negative control reactions scored positive within 24 hours. Positive controls were seeded with blood from sheep clinically affected with BSE, and serial dilutions of the reference BSE-infected sheep brain homogenate (BSB/7/10).
Statistical analysis
Infectivity titers (ID50/mL WB) from the TgBov mouse bioassay (terminal time point vs the penultimate time point tested) were compared using a nonparametric Wilcoxon matched-pairs signed-rank test. Data for donor sheep N218 were omitted from the analysis because 0 infectivity at the terminal time point was deemed an anomalous result. The relationship between infectivity titer and limit of detection (LoD) in PMCA was inferred by the nonparametric Spearman correlation (2-tailed). Statistical analyses were performed in GraphPad Prism version 10.2.3 (GraphPad Software Inc, CA), with P values <.05 considered significant.
Mouse experiments were approved by the local ethics committee of the National Veterinary School of Toulouse, France. Archived sheep blood samples were obtained during a previous project that was approved by the animal welfare and ethical review committee of The Roslin Institute and licensed by the United Kingdom Home Office (Project license numbers: 30/2282, 60/4143, and 70/8595).
Results
Comparison of the analytical sensitivity of in vitro assays
To compare the analytical sensitivity of the 3 different assays, and their cross-species reactivity, each assay was tested on reference brain homogenates from BSE-infected sheep (BSB/7/10) and a patient with vCJD (NIBSC reference: NHBY0/0003; Table 1). The results for mb-PMCA and RT-QuIC were reported previously,35 but are included here for comparison with capture-PMCA. For BSE-infected sheep brain homogenate, mb-PMCA and capture-PMCA showed similar analytical sensitivities (limits of detection at 10−9 to 10−10 dilution of brain homogenate), which are greater than that of RT-QuIC (LoD at 10−7 to 10−8 dilution). mb-PMCA and RT-QuIC were marginally less sensitive in detecting PrPSc in the reference vCJD brain homogenate (limits of detection at dilutions of 10−9 and 10−6, respectively), whereas capture PMCA showed similar levels of sensitivity on vCJD-infected and BSE-infected brain homogenates.
Comparison of the analytical sensitivity of in vitro assays on reference brain homogenates from BSE-infected sheep and a patient with vCJD
| Assay . | Seed . | Dilution of prion-infected brain homogenate . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10−1 . | 10−2 . | 10−3 . | 10−4 . | 10−5 . | 10−6 . | 10−7 . | 10−8 . | 10−9 . | 10−10 . | 10−11 . | ||
| RT-QuIC∗ | vCJD | ND | ND | ND | 1 | 0.7 | 0.1 | 0 | 0 | 0 | 0.1 | ND |
| ShBSE | ND | ND | ND | 1 | 1 | 1 | 0.7 | 0.1 | 0 | 0 | ND | |
| mb-PMCA† | vCJD | ND | ND | ND | 1 | 1 | 1 | 1 | 0.3 | 0.1 | 0 | ND |
| ShBSE | ND | ND | ND | ND | 1 | 1 | 1 | 1 | 0.8 | 0.2 | 0 | |
| capture-PMCA‡ | vCJD | ND | ND | ND | ND | ND | 1 | 1 | 1 | 1 | 0.2 | 0.2 |
| ShBSE | ND | ND | ND | ND | 1 | 1 | 1 | 1 | 1 | 0.3 | ND | |
| Assay . | Seed . | Dilution of prion-infected brain homogenate . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10−1 . | 10−2 . | 10−3 . | 10−4 . | 10−5 . | 10−6 . | 10−7 . | 10−8 . | 10−9 . | 10−10 . | 10−11 . | ||
| RT-QuIC∗ | vCJD | ND | ND | ND | 1 | 0.7 | 0.1 | 0 | 0 | 0 | 0.1 | ND |
| ShBSE | ND | ND | ND | 1 | 1 | 1 | 0.7 | 0.1 | 0 | 0 | ND | |
| mb-PMCA† | vCJD | ND | ND | ND | 1 | 1 | 1 | 1 | 0.3 | 0.1 | 0 | ND |
| ShBSE | ND | ND | ND | ND | 1 | 1 | 1 | 1 | 0.8 | 0.2 | 0 | |
| capture-PMCA‡ | vCJD | ND | ND | ND | ND | ND | 1 | 1 | 1 | 1 | 0.2 | 0.2 |
| ShBSE | ND | ND | ND | ND | 1 | 1 | 1 | 1 | 1 | 0.3 | ND | |
Data for RT-QuIC and mb-PMCA were published previously.
ND, not determined; ShBSE, sheep BSE; vCJD, variant Creutzfeldt-Jakob disease.
RT-QuIC data represent the mean proportion of positive replicates from 3 independent experiments.
mb-PMCA data represent the mean proportion of positive replicates from 2 or 3 independent experiments.
capture-PMCA data represent the mean proportion of positive replicates from 6 or 8 independent experiments for vCJD and ShBSE seeds spiked in human plasma or sheep buffy coat, respectively.
Comparison of tests on blinded panels of prion-infected blood samples
Blinded panels of buffy coat or whole-blood samples collected from identical sheep and time points were supplied to the laboratories involved for testing with the assays. Each panel included longitudinal series of samples from individual BSE-infected donor and recipient sheep, and mock-infected negative controls, collected throughout preclinical and clinical stages of infection. Additional samples collected from donors at the time that blood was collected for transfusion (10 mpi) were included to compare test results with the outcomes of transfusion in recipients. The total number of samples analyzed by WB IOME RT-QuIC and mb-PMCA was 139, whereas the total number analyzed by capture-PMCA was 128 (because of a moratorium on prion research in France that interrupted the study).
For all blinded samples, mb-PMCA gave positive results on 77 of 102 samples (76%) from BSE-infected sheep, compared with 68 of 94 (72%) and 55 of 102 (54%) identified by capture-PMCA and WB IOME RT-QuIC, respectively (Table 2; supplemental Figures 2-4). For blood samples collected during the clinical phase of disease (n = 18), all were correctly identified by mb-PMCA and capture-PMCA (diagnostic sensitivity of 100%), whereas 11 of 18 tested positive by WB IOME RT-QuIC (diagnostic sensitivity of 61%). None of the assays generated false positives when tested on samples from mock-infected negative control sheep (n = 12), equating to a diagnostic specificity of 100% (95% confidence interval, 90.5-100) for all 3 blood tests.
Summary of results from blinded panels
| Assay . | Samples from BSE-infected sheep, n = 102 or 94∗ . | Samples from mock-infected sheep, n = 37 or 34∗ . | ||
|---|---|---|---|---|
| Positive test result . | Negative test result . | Positive test result . | Negative test result . | |
| mb-PMCA | 77 | 25 | 0 | 37 |
| capture-PMCA | 68 | 26 | 0 | 34 |
| WB IOME RT-QuIC | 55 | 47 | 0 | 37 |
| Assay . | Samples from BSE-infected sheep, n = 102 or 94∗ . | Samples from mock-infected sheep, n = 37 or 34∗ . | ||
|---|---|---|---|---|
| Positive test result . | Negative test result . | Positive test result . | Negative test result . | |
| mb-PMCA | 77 | 25 | 0 | 37 |
| capture-PMCA | 68 | 26 | 0 | 34 |
| WB IOME RT-QuIC | 55 | 47 | 0 | 37 |
A smaller total number of samples was tested by capture-PMCA because the laboratory was unable to test the second “mini panel” of 11 samples.
The positive predictive values for all 3 assays are 100%; negative predictive values for mb-PMCA and capture-PMCA are also 100%, and 84.1% for the RT-QuIC assay (supplemental Table 1). Receiver operator characteristic analysis was used to retrospectively assess the effect of varying threshold fluorescence on RT-QuIC assay performance and evaluate the overall accuracy of the assay.48 The analysis indicated an area under the curve of 0.82 (95% confidence interval, 0.69-0.95) and an optimal threshold of 60% maximum ThT fluorescence reading for the plate (Fmax) to maximize sensitivity without compromising specificity (supplemental Figure 5).
PrPSc detection in blood over the time course of infection
From the results of the blinded study, we were able to compare the earliest preclinical time points at which infected sheep were identified by the different assays, and the consistency of test results on samples from individual infected sheep over the course of infection.
Each of the in vitro assays was more effective at detecting PrPSc in blood samples from recipient sheep, compared with donor sheep, during preclinical time points spanning 6 to 18 mpi. For donors, the onset of detection of PrPSc in the blood was variable, ranging from 6 to 18 mpi (equating to 10-27 months before clinical onset) for mb-PMCA and capture-PMCA. For WB IOME RT-QuIC, the earliest time point at which blood samples from infected donors tested positive ranged from 10 to 18 mpi (equating to 2-22 months before clinical onset; Table 3; supplemental Figures 2-4). From 18 mpi until clinical onset, all blood samples tested positive with each of the 3 assays.
Summary of testing with in vitro assays on longitudinal series of blood samples from BSE-infected donor sheep
| Assay . | No. of positive samples/no. of samples tested (% positive samples) . | |||||
|---|---|---|---|---|---|---|
| TP1 2 mpi . | TP2 6 mpi . | TP3 10 mpi . | TP4 18 mpi . | TP5 27-28 mpi . | TP6 (terminal) 18-39 mpi . | |
| mb-PMCA | 0/9 (0%) | 4/9 (44%) | 6/8 (75%) | 8/8 (100%) | 3/3 (100%) | 9/9 (100%) |
| capture-PMCA | 0/9 (0%) | 3/9 (33%) | ND | 8/8 (100%) | 3/3 (100%) | 9/9 (100%) |
| WB IOME RT-QuIC | 0/9 (0%) | 0/9 (0%) | 3/8 (38%) | 8/8 (100%) | 3/3 (100%) | 5/9 (56%) |
| Assay . | No. of positive samples/no. of samples tested (% positive samples) . | |||||
|---|---|---|---|---|---|---|
| TP1 2 mpi . | TP2 6 mpi . | TP3 10 mpi . | TP4 18 mpi . | TP5 27-28 mpi . | TP6 (terminal) 18-39 mpi . | |
| mb-PMCA | 0/9 (0%) | 4/9 (44%) | 6/8 (75%) | 8/8 (100%) | 3/3 (100%) | 9/9 (100%) |
| capture-PMCA | 0/9 (0%) | 3/9 (33%) | ND | 8/8 (100%) | 3/3 (100%) | 9/9 (100%) |
| WB IOME RT-QuIC | 0/9 (0%) | 0/9 (0%) | 3/8 (38%) | 8/8 (100%) | 3/3 (100%) | 5/9 (56%) |
Please note time points TP1-TP5 are preclinical; clinical signs of BSE were evident only at TP6 (terminal).
ND, not determined; TP, time point.
In contrast, almost all samples from recipient animals tested positive from the earliest time point sampled (6 mpi; Table 4; supplemental Figures 2-4), with 9 of 10 (90%), 8 of 10 (80%), and 7 of 10 (70%) samples at this time point testing positive by mb-PMCA, capture-PMCA, and WB IOME RT-QuIC, respectively. This equates to detection up to 26 months before clinical onset for all 3 assays. All samples from subsequent time points (9 mpi onward) tested positive in the PMCA assays, and the majority (≥80%) also tested positive by WB IOME RT-QuIC until the terminal stage of infection.
Summary of testing with in vitro assays on longitudinal series of blood samples from BSE-infected recipient sheep
| Assay . | No. of positive samples/no. of samples tested (% positive samples) . | |||||
|---|---|---|---|---|---|---|
| TP1 6 mpi . | TP2 9 mpi . | TP3 12 mpi . | TP4 18-25∗ mpi . | TP5 32 mpi . | TP6 (terminal)15-36 mpi . | |
| mb-PMCA | 9/10 (90%) | 10/10 (100%) | 10/10 (100%) | 3/4 (75%) | 1/1 (100%) | 9/9 (100%) |
| capture-PMCA | 8/10 (80%) | 10/10 (100%) | 10/10 (100%) | 3/4 (75%) | 1/1 (100%) | 9/9 (100%) |
| WB IOME RT-QuIC | 7/10 (70%) | 8/10 (80%) | 9/10 (90%) | 3/4 (75%) | 1/1 (100%) | 6/9 (67%) |
| Assay . | No. of positive samples/no. of samples tested (% positive samples) . | |||||
|---|---|---|---|---|---|---|
| TP1 6 mpi . | TP2 9 mpi . | TP3 12 mpi . | TP4 18-25∗ mpi . | TP5 32 mpi . | TP6 (terminal)15-36 mpi . | |
| mb-PMCA | 9/10 (90%) | 10/10 (100%) | 10/10 (100%) | 3/4 (75%) | 1/1 (100%) | 9/9 (100%) |
| capture-PMCA | 8/10 (80%) | 10/10 (100%) | 10/10 (100%) | 3/4 (75%) | 1/1 (100%) | 9/9 (100%) |
| WB IOME RT-QuIC | 7/10 (70%) | 8/10 (80%) | 9/10 (90%) | 3/4 (75%) | 1/1 (100%) | 6/9 (67%) |
Please note time points TP1-TP5 are preclinical: clinical signs of BSE were evident only at TP6 (terminal).
Twenty-five mpi sample from sheep P453 tested negative by all 3 in vitro assays, despite earlier and subsequent time points testing positive (also see supplemental Figures 2-4); the reason for this apparently anomalous result could not be determined.
For both PMCA assays, once a preclinical blood sample from an individual sheep tested positive, samples from the same sheep at all subsequent time points were also positive (with rare exceptions). However, a significant proportion of blood samples (7/18) from the terminal time point (when sheep were showing clinical signs of BSE) tested negative with WB IOME RT-QuIC, despite samples from the same sheep at late preclinical stages testing positive (Tables 3 and 4). Given the lower analytical sensitivity of RT-QuIC compared with the 2 PMCA assays, the most likely explanation for this is a decrease in PrPSc levels in blood during the clinical phase of infection.
Comparison of blood infectivity titers and assay results over the time course of infection
Previous studies have shown that PrPSc levels in tissues/body fluids and titers of infectious prions are not always closely correlated.49 Therefore, buffy coat samples from 3 BSE-infected donors, 3 BSE-infected recipients, and 1 negative control sheep included in the blinded panel (n = 33) were intracerebrally inoculated into TgBov mice42,43 to determine infectivity titers. TgBov mice have been used as a sensitive bioassay host for blood samples from BSE-infected sheep and patients with vCJD.18
There was a significant positive correlation between bioassay results and in vitro test results, in that samples that transmitted infection to TgBov mice gave positive results in ≥1 of the in vitro assays (Table 5). Two samples from donor N218 at early time points (6 and 10 mpi) failed to transmit infection to mice and also tested negative in all 3 in vitro assays. In contrast, 6 mpi samples from donor N236, which also failed to transmit infection to mice, tested positive by mb-PMCA and capture-PMCA (Table 5), suggesting that the PMCA assays are marginally more sensitive than bioassay in identifying samples from infected animals.
Comparison of blood infectivity titers measured by TgBov mouse bioassay with results of in vitro blood tests
| Sheep ID . | Status . | Time point, mpi . | No. of positive mice/no. of mice inoculated∗ . | Log10 infectivity titer ID50/mL WB (95% CI)† . | Result of testing blinded panels with in vitro assays . | LoD by mb-PMCA‡ . | ||
|---|---|---|---|---|---|---|---|---|
| mb-PMCA . | capture-PMCA . | WB IOME RT-QuIC . | ||||||
| N236 | Donor | 6 | 0/12 | 0 (0-2.5) | + | + | − | 10−1 |
| 10 | 9/12 | 3.5 (3.1-3.9) | + | ND | + | 10−4 | ||
| 12 | 11/12 | 3.5 (3.1-3.9) | ND | ND | ND | 10−4 | ||
| 18/terminal | 8/12 | 3.3 (2.9-3.7) | + | + | + | 10−4 | ||
| N218 | Donor | 6 | 0/12 | 0 (0-2.4) | − | − | − | NA |
| 10 | 0/11 | 0 (0-2.5) | − | ND | − | NA | ||
| 18 | 11/12 | 3.6 (3.2-4.0) | + | + | + | 10−4 | ||
| 25 | 8/12 | 3.0 (2.7-3.4) | ND | ND | ND | 10−4 | ||
| 28/terminal | 0/12 | 0 (0-2.4)§ | + | + | + | 10−4 | ||
| N245 | Donor | 6 | 0/12 | 0 (0-2.4) | +/− | − | − | ND |
| 10 | 9/12 | 2.8 (2.6-3.2) | + | ND | + | 10−2 | ||
| 18 | 12/12 | 3.9 (3.5-4.3) | + | + | + | 10−5 | ||
| 28 | 10/12 | 3.6 (3.2-4.0) | + | + | + | 10−5 | ||
| 32/terminal | 8/12 | 2.8 (2.5-3.1) | + | + | + | 10−4 | ||
| P480 | Recipient | 6 | 11/12 | 3.3 (2.9-3.7) | + | + | + | 10−5 |
| 9 | 11/12 | 3.8 (3.4-4.2) | + | + | + | 10−4 | ||
| 12 | 12/12 | 3.8 (3.4-4.2) | + | + | + | 10−4 | ||
| 16/terminal | 11/12 | 3.4 (3.1-3.8) | + | + | + | 10−3 | ||
| M246 | Recipient | 6 | 10/12 | 2.9 (2.6-3.2) | + | + | + | 10−3 |
| 9 | 9/12 | 3.0 (2.7-3.4) | + | + | + | 10−4 | ||
| 12 | 10/12 | 3.3 (2.9-3.7) | + | + | + | 10−3 | ||
| 15 | 11/11 | 3.2 (2.9-3.6) | ND | ND | ND | 10−4 | ||
| 19/terminal | 5/12 | 2.7 (2.5-3.0) | + | + | + | 10−3 | ||
| M490 | Recipient | 6 | 6/11 | 2.7 (2.4-3.0) | + | + | + | 10−3 |
| 9 | 8/12 | 3.1 (2.8-3.6) | + | + | + | 10−4 | ||
| 15 | 8/12 | 3.0 (2.7-3.3) | ND | ND | ND | 10−3 | ||
| 21 | 6/12 | 2.8 (2.5-3.1) | ND | ND | ND | 10−4 | ||
| 26/terminal | 3/12 | 2.4 (2.0-2.7) | + | + | + | 10−4 | ||
| N239 | Negative control | 6 | 0/12 | 0 (0-2.3) | − | − | − | NA |
| 10 | 0/12 | 0 (0-2.4) | − | ND | − | NA | ||
| 12 | 0/12 | 0 (0-2.3) | ND | ND | ND | NA | ||
| 27 | 0/12 | 0 (0-2.3) | − | − | − | NA | ||
| 34 | 0/12 | 0 (0-2.3) | − | − | − | NA | ||
| Sheep ID . | Status . | Time point, mpi . | No. of positive mice/no. of mice inoculated∗ . | Log10 infectivity titer ID50/mL WB (95% CI)† . | Result of testing blinded panels with in vitro assays . | LoD by mb-PMCA‡ . | ||
|---|---|---|---|---|---|---|---|---|
| mb-PMCA . | capture-PMCA . | WB IOME RT-QuIC . | ||||||
| N236 | Donor | 6 | 0/12 | 0 (0-2.5) | + | + | − | 10−1 |
| 10 | 9/12 | 3.5 (3.1-3.9) | + | ND | + | 10−4 | ||
| 12 | 11/12 | 3.5 (3.1-3.9) | ND | ND | ND | 10−4 | ||
| 18/terminal | 8/12 | 3.3 (2.9-3.7) | + | + | + | 10−4 | ||
| N218 | Donor | 6 | 0/12 | 0 (0-2.4) | − | − | − | NA |
| 10 | 0/11 | 0 (0-2.5) | − | ND | − | NA | ||
| 18 | 11/12 | 3.6 (3.2-4.0) | + | + | + | 10−4 | ||
| 25 | 8/12 | 3.0 (2.7-3.4) | ND | ND | ND | 10−4 | ||
| 28/terminal | 0/12 | 0 (0-2.4)§ | + | + | + | 10−4 | ||
| N245 | Donor | 6 | 0/12 | 0 (0-2.4) | +/− | − | − | ND |
| 10 | 9/12 | 2.8 (2.6-3.2) | + | ND | + | 10−2 | ||
| 18 | 12/12 | 3.9 (3.5-4.3) | + | + | + | 10−5 | ||
| 28 | 10/12 | 3.6 (3.2-4.0) | + | + | + | 10−5 | ||
| 32/terminal | 8/12 | 2.8 (2.5-3.1) | + | + | + | 10−4 | ||
| P480 | Recipient | 6 | 11/12 | 3.3 (2.9-3.7) | + | + | + | 10−5 |
| 9 | 11/12 | 3.8 (3.4-4.2) | + | + | + | 10−4 | ||
| 12 | 12/12 | 3.8 (3.4-4.2) | + | + | + | 10−4 | ||
| 16/terminal | 11/12 | 3.4 (3.1-3.8) | + | + | + | 10−3 | ||
| M246 | Recipient | 6 | 10/12 | 2.9 (2.6-3.2) | + | + | + | 10−3 |
| 9 | 9/12 | 3.0 (2.7-3.4) | + | + | + | 10−4 | ||
| 12 | 10/12 | 3.3 (2.9-3.7) | + | + | + | 10−3 | ||
| 15 | 11/11 | 3.2 (2.9-3.6) | ND | ND | ND | 10−4 | ||
| 19/terminal | 5/12 | 2.7 (2.5-3.0) | + | + | + | 10−3 | ||
| M490 | Recipient | 6 | 6/11 | 2.7 (2.4-3.0) | + | + | + | 10−3 |
| 9 | 8/12 | 3.1 (2.8-3.6) | + | + | + | 10−4 | ||
| 15 | 8/12 | 3.0 (2.7-3.3) | ND | ND | ND | 10−3 | ||
| 21 | 6/12 | 2.8 (2.5-3.1) | ND | ND | ND | 10−4 | ||
| 26/terminal | 3/12 | 2.4 (2.0-2.7) | + | + | + | 10−4 | ||
| N239 | Negative control | 6 | 0/12 | 0 (0-2.3) | − | − | − | NA |
| 10 | 0/12 | 0 (0-2.4) | − | ND | − | NA | ||
| 12 | 0/12 | 0 (0-2.3) | ND | ND | ND | NA | ||
| 27 | 0/12 | 0 (0-2.3) | − | − | − | NA | ||
| 34 | 0/12 | 0 (0-2.3) | − | − | − | NA | ||
CI, confidence interval; NA, not applicable; +, positive; +/−, weak positive; −, negative.
Number of TgBov mice that tested positive/total number of mice inoculated.
Log10 infectivity titer (equivalent to Log10 ID50/mL WB), as determined by intracerebral inoculation in TgBov mice. 95% confidence intervals are shown in parentheses.
LoD determined independently by testing a dilution series of corresponding sample by mb-PMCA.
Terminal blood sample (28 mpi) from donor N218 did not transmit infection to mice, despite testing positive in all 3 in vitro assays, and positive transmissions from samples at earlier time points (18 mpi and 25 mpi); the reason for this apparently anomalous result could not be determined.
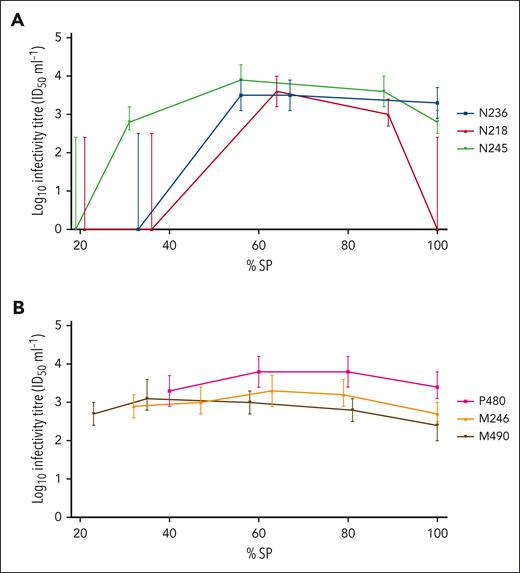
The onset of detection of infectivity in blood was more variable in donor than recipient sheep, reflecting results from the 3 assays. Once infectivity was detected in blood, the titers in individual sheep remained relatively constant during the course of infection (Table 5). In several animals, a slight decrease in titer was observed at the terminal (clinical) time point (Figure 1); however, this was not statistically significant (P = .06, Wilcoxon matched-pairs signed-rank test).
Time course of infectivity titer in representative BSE-infected donor and recipient sheep, as determined by mouse bioassay. Infectivity titers (ID50/mL WB), as determined by end point titration in TgBov mice, were plotted according to percentage survival period (% SP), for 3 BSE-infected donor sheep N236, N218, and N245 (colored blue, red and green, respectively) (A); and 3 BSE-infected recipients P480, M246, and M490 (colored pink, orange and black, respectively) (B). Error bars represent 95% confidence intervals.
Time course of infectivity titer in representative BSE-infected donor and recipient sheep, as determined by mouse bioassay. Infectivity titers (ID50/mL WB), as determined by end point titration in TgBov mice, were plotted according to percentage survival period (% SP), for 3 BSE-infected donor sheep N236, N218, and N245 (colored blue, red and green, respectively) (A); and 3 BSE-infected recipients P480, M246, and M490 (colored pink, orange and black, respectively) (B). Error bars represent 95% confidence intervals.
We also used mb-PMCA to perform end point dilution of all blood samples that transmitted infection (ie, samples that transmitted infection to at least 1 inoculated mouse within that cohort), to determine the LoD as a proxy for PrPSc concentration (Table 5). This analysis indicated a moderate positive correlation between infectivity titer in TgBov mouse bioassay and LoD in mb-PMCA (Spearman coefficient, r = 0.51, P ≤ .01; supplemental Figure 6).
Relationship between assay results and infectivity for mice and sheep
Because there was detailed retrospective data on which sheep were infected with BSE, we were able to compare the assay results on blood samples from donor sheep at the time of blood collection for transfusion (10 mpi), with the outcomes of transfusion in the respective recipients of different blood components. Thus, a subset of samples in the blinded panels were from BSE-infected donors (at 10 mpi) that either transmitted BSE to ≥1 transfusion recipients (“transmitters”; n = 12) or did not transmit BSE to any recipients (“nontransmitters”; n = 8); some of these samples were also inoculated intracerebrally into TgBov mice. The results are summarized in Table 6, and details of samples are given in supplemental Figures 2-4.
Comparison of transfusion transmission outcomes in sheep with results of in vitro assays and blood infectivity titers
| Results from sheep transfusion experiments . | Results of in vitro assays . | Results of mouse bioassay∗ . | ||||||
|---|---|---|---|---|---|---|---|---|
| Donor sheep ID† . | Transfusion outcome . | No. of recipients infected/no. of recipients transfused‡ . | Donor survival period, mpi§ . | mb-PMCA . | capture-PMCA . | WB IOME RT-QuIC . | No. of positive mice/no. of mice inoculated . | Log10 infectivity titers/mL WB (95% CI) . |
| N157 | T | 2/7 | 20 | +/− | + | − | ND | ND |
| N231 | T | 1/7 | 20 | +/− | + | − | ND | ND |
| N251 | T | 5/5 | 18 | + | + | + | ND | ND |
| N261 | T | 6/7 | 19 | + | + | + | ND | ND |
| N257 | T | 5/5 | 20 | + | ND | + | ND | ND |
| N232 | T | 5/7 | 21 | + | ND | − | 1/12 | 2.2 (1.7-2.6) |
| N236 | T | 5/5 | 12 | + | ND | + | 9/12 | 3.5 (3.1-3.9)|| |
| N226 | T | 1/7 | 28 | − | ND | − | ND | ND |
| N180 | T | 3/7 | 27 | + | ND | − | 0/12 | 0 (0-2.3) |
| N218 | T | 1/5 | 28 | − | ND | − | 0/11 | 0 (0-2.5)|| |
| N204 | T | 2/7 | 37 | + | ND | − | 0/12 | 0 (0-2.3) |
| N245 | T | 1/4 | 32 | + | ND | + | 9/12 | 2.8 (2.6-3.2)|| |
| N196 | NT | 0/6 | 20 | − | − | − | 0/12 | 0 (0-2.3) |
| N258 | NT | 0/7 | 31 | − | − | − | ND | ND |
| N188 | NT | 0/5 | 22 | − | − | − | ND | ND |
| N220 | NT | 0/5 | 26 | − | − | − | 0/12 | 0 (0-2.3) |
| N228 | NT | 0/4 | 31 | − | − | − | ND | ND |
| N234 | NT | 0/5 | 36 | − | − | − | 1/12 | 2.2 (1.6-2.6) |
| N246 | NT | 0/7 | 46 | − | − | − | ND | ND |
| N161 | NT | 0/7 | 39 | +/− | − | − | ND | ND |
| N239 | Negative control | NA | NA | − | ND | − | 0/12 | 0 (0-2.4) |
| N170 | Negative control | NA | NA | − | ND | − | ND | ND |
| N214 | Negative control | NA | NA | − | ND | − | ND | ND |
| N173 | Negative control | NA | NA | − | − | − | ND | ND |
| N217 | Negative control | NA | NA | − | − | − | ND | ND |
| N256 | Negative control | NA | NA | − | − | − | ND | ND |
| N155 | Negative control | NA | NA | − | − | − | ND | ND |
| N209 | Negative control | NA | NA | − | − | − | ND | ND |
| N152 | Negative control | NA | NA | − | − | − | ND | ND |
| Results from sheep transfusion experiments . | Results of in vitro assays . | Results of mouse bioassay∗ . | ||||||
|---|---|---|---|---|---|---|---|---|
| Donor sheep ID† . | Transfusion outcome . | No. of recipients infected/no. of recipients transfused‡ . | Donor survival period, mpi§ . | mb-PMCA . | capture-PMCA . | WB IOME RT-QuIC . | No. of positive mice/no. of mice inoculated . | Log10 infectivity titers/mL WB (95% CI) . |
| N157 | T | 2/7 | 20 | +/− | + | − | ND | ND |
| N231 | T | 1/7 | 20 | +/− | + | − | ND | ND |
| N251 | T | 5/5 | 18 | + | + | + | ND | ND |
| N261 | T | 6/7 | 19 | + | + | + | ND | ND |
| N257 | T | 5/5 | 20 | + | ND | + | ND | ND |
| N232 | T | 5/7 | 21 | + | ND | − | 1/12 | 2.2 (1.7-2.6) |
| N236 | T | 5/5 | 12 | + | ND | + | 9/12 | 3.5 (3.1-3.9)|| |
| N226 | T | 1/7 | 28 | − | ND | − | ND | ND |
| N180 | T | 3/7 | 27 | + | ND | − | 0/12 | 0 (0-2.3) |
| N218 | T | 1/5 | 28 | − | ND | − | 0/11 | 0 (0-2.5)|| |
| N204 | T | 2/7 | 37 | + | ND | − | 0/12 | 0 (0-2.3) |
| N245 | T | 1/4 | 32 | + | ND | + | 9/12 | 2.8 (2.6-3.2)|| |
| N196 | NT | 0/6 | 20 | − | − | − | 0/12 | 0 (0-2.3) |
| N258 | NT | 0/7 | 31 | − | − | − | ND | ND |
| N188 | NT | 0/5 | 22 | − | − | − | ND | ND |
| N220 | NT | 0/5 | 26 | − | − | − | 0/12 | 0 (0-2.3) |
| N228 | NT | 0/4 | 31 | − | − | − | ND | ND |
| N234 | NT | 0/5 | 36 | − | − | − | 1/12 | 2.2 (1.6-2.6) |
| N246 | NT | 0/7 | 46 | − | − | − | ND | ND |
| N161 | NT | 0/7 | 39 | +/− | − | − | ND | ND |
| N239 | Negative control | NA | NA | − | ND | − | 0/12 | 0 (0-2.4) |
| N170 | Negative control | NA | NA | − | ND | − | ND | ND |
| N214 | Negative control | NA | NA | − | ND | − | ND | ND |
| N173 | Negative control | NA | NA | − | − | − | ND | ND |
| N217 | Negative control | NA | NA | − | − | − | ND | ND |
| N256 | Negative control | NA | NA | − | − | − | ND | ND |
| N155 | Negative control | NA | NA | − | − | − | ND | ND |
| N209 | Negative control | NA | NA | − | − | − | ND | ND |
| N152 | Negative control | NA | NA | − | − | − | ND | ND |
NT, no transmission; T, transmission (transmission of infection from BSE-infected donor to ≥1 transfusion recipients); +, positive; +/−, weak positive; −, negative.
TgBov mouse bioassay (intracerebral), log10 infectivity titer (ID50/mL WB).
Samples from N257, N232, N236, N226, N180, N218, N204, and N245 (underlined) included in the longitudinal blood series (see supplemental Figures 2-4).
Number of BSE-infected recipient sheep (clinically and/or pathologically positive)/the number of recipients of blood components from a single BSE-infected donor.
Survival period, interval between the date of infection, and the date of euthanasia (on reaching humane clinical end points).
Titers from 10 mpi time point in longitudinal blood series (also shown in Table 5).
Almost all blood samples that tested positive in mb-PMCA, capture-PMCA, and WB IOME RT-QuIC were from donors that transmitted infection to transfusion recipients. Samples from nontransmitters were negative with all 3 assays, apart from 1 that was weakly positive in mb-PMCA (Table 6). mb-PMCA gave positive results on a higher proportion of samples from transmitters (10/12, including 2 weak positives) than WB IOME RT-QuIC (5/12). The majority of samples that tested positive by both mb-PMCA and WB IOME RT-QuIC (4/5) came from donor sheep with the highest rates of transmission to recipients (86%-100% recipients infected).
Of 6 buffy coat samples from transmitters, 3 also transmitted infection when inoculated intracerebrally into TgBov mice (Table 6). The 2 samples with the highest estimated titers (103.5 and 102.8 ID50/mL WB, respectively) tested positive by both mb-PMCA and WB IOME RT-QuIC, whereas the third sample (estimated titer 102.2 ID50/mL WB) tested positive by mb-PMCA only. Of 3 samples from nontransmitters, 1 also transmitted infection to TgBov mice (1/12 mice infected; estimated titer 102.2 ID50/mL WB) but tested negative by mb-PMCA and WB IOME RT-QuIC. These slight discrepancies probably reflect that infectivity in the blood samples was nonuniformly distributed, and close to the LoD of the bioassay.
Collectively, these results demonstrate that samples from sheep whose blood contained sufficient infectivity to transmit infection by transfusion were also almost exclusively those that tested positive by PMCA and WB IOME RT-QuIC, and transmitted BSE to TgBov mice for estimation of infectivity titers. This provides further evidence for a positive correlation between PrPSc levels and prion infectivity in blood.
Discussion
This study aimed to evaluate and compare the performance of 3 in vitro PrPSc amplification assays for detection of preclinical prion infection, using blinded panels of blood samples from BSE-infected sheep, an established animal model for vCJD. The 2 PMCA assays tested (mb-PMCA and capture-PMCA) had roughly equivalent analytical sensitivities for PrPSc detection, which were ∼2 orders of magnitude (log10) greater than that of WB IOME RT-QuIC. Accordingly, when applied to the blinded panels, diagnostic sensitivity of mb-PMCA and capture-PMCA was 100% for samples collected during the clinical phase of infection, compared with 61% for WB IOME RT-QuIC. The PMCA assays were also able to identify a higher proportion of samples from infected animals at earlier preclinical stages than WB IOME RT-QuIC. All 3 assays correctly identified the samples from negative control animals, a very important consideration when considering deployment of a vCJD test for screening and/or diagnosis. However, further analysis using a larger noninfected cohort would be advisable to confirm 100% diagnostic specificity.
The higher diagnostic sensitivity of PMCA assays compared with WB IOME RT-QuIC would suggest that the former are more appropriate for detection of preclinical prion infection. However, RT-QuIC has advantages over PMCA, which may make it more adaptable for routine use in diagnostic laboratories. It is a simpler, more rapid procedure, using recPrP rather than uninfected mouse brain homogenates as substrate, and does not amplify infectivity along with PrPSc. The diagnostic sensitivity of WB IOME RT-QuIC could potentially be improved by further technical optimization, for example by adaptation to buffy coat samples, which contain higher levels of infectivity and PrPSc than WB.36
Although the diagnostic sensitivity of WB IOME RT-QuIC was 61% when applied to clinical blood samples, the assay gave positive results on almost all samples (94%) from later preclinical time points (≥18 mpi) in the same sheep. This suggests that PrPSc concentrations in blood decrease during clinical disease, which is consistent with earlier observations of significantly longer lag times in RT-QuIC reactions for blood samples from clinical compared with preclinical time points.35 Thus, the diagnostic sensitivity of WB IOME RT-QuIC underestimates its ability to detect preclinical infection. Parallel estimation of infectivity in longitudinal buffy coat sample series by TgBov mouse bioassay showed that blood titers remained relatively stable throughout infection but decreased at the clinical phase in some sheep. The reason for the decrease in PrPSc levels and infectivity titers during clinical disease is not clear. Similar findings have not been widely reported in other animal models of prion disease, although a decrease in blood infectivity titers was also found at clinical end points in CD-1 mice infected intracerebrally with Rocky Mountains Laboratory (RML) scrapie.49
Detection of PrPSc and infectivity in blood across the time course of infection differed substantially between donor and recipient sheep, with relatively later and more variable onset of detection in orally infected donors. Similar results were previously reported in an unblinded analysis of blood samples from the same archive by mb-PMCA.36 A number of factors may contribute to this difference, including infectious dose and source of infectivity (ie, brain vs blood), but comparison with similar studies suggests that route of infection could be important. Cynomolgus macaques infected with vCJD by a combination of intravenous and intraperitoneal injection showed PrPSc detection in blood from 2 mpi by PMCA,25,50 which is similar to our results in recipient sheep infected by intravenous transfusion. In contrast, in deer naturally infected with chronic wasting disease (CWD), only 53% of samples from individuals in the early preclinical phase of infection tested positive by PMCA,51 comparable with the results we observed in orally infected donors. Given that the majority of human exposure to BSE is likely to have been via the oral route, our results suggest that the route and method of infection must be considered when extrapolating from detection of preclinical infection in animal models to humans.
Comparison of in vitro assay results with infectivity titers in TgBov mice and outcomes of blood transfusion in sheep provide further evidence for a correlation between PrPSc concentration and the level of infectivity in blood. In general, samples that tested positive by PMCA and/or WB IOME RT-QuIC also transmitted infection to TgBov mice. For samples assayed in parallel, there was a modest positive correlation between infectivity titers and the limit of PrPSc detection by end point dilution in mb-PMCA (a proxy for PrPSc concentration). In addition, the majority of blood samples that transmitted prion infection by transfusion in sheep tested positive by mb-PMCA, whereas almost all samples that were not infectious for sheep tested negative.
All 3 assays showed almost equivalent analytical sensitivities when tested on reference vCJD-infected human and BSE-infected sheep brain homogenates,35 suggesting that they should be applicable to either sheep or human blood samples with only minor modification. Application of these results to human medicine also depends on the extent to which titers of prion infectivity/PrPSc in the blood of BSE-infected sheep reflect those in patients with vCJD. Previous studies suggest that PrPSc levels in the lymphoid tissues of patients with vCJD52 are comparable with those found in BSE-infected sheep,53 which is reflected in estimates of infectivity in the same tissues by TgBov mouse bioassay.54 Estimation of PrPSc levels by end point dilution in PMCA indicate a similar LoD for buffy coat samples from symptomatic vCJD cases38 to that reported here in sheep clinically affected with BSE (10−3 to 10−5 sample dilution). The blood infectivity titers estimated by infection of TgBov mice were higher (2-3 log difference) than those previously reported after inoculation of the same samples in TgShpXI mice.36 However, it is not possible to directly compare titers obtained in different bioassay hosts, and the discrepancy is most likely because of greater sensitivity of TgBov mice to BSE. After intracerebral inoculation of leukocytes from 1 patient with vCJD in TgBov mice, the estimated titer was 2.2 ID/mL.18 This may suggest that infectivity titers are much lower in the blood of patients with vCJD than in BSE-infected sheep, but bioassay of larger numbers of samples from patients with vCJD is necessary for robust comparison.
In summary, we have shown highly sensitive and specific detection of prion-infected blood samples from early preclinical time points through to clinical disease, using 3 in vitro PrPSc amplification assays. PMCA assays had greater diagnostic sensitivity than WB IOME RT-QuIC but may be less amenable to routine diagnosis/screening. Detection of PrPSc in blood reflected the levels of infectivity present, suggesting it could be used for estimation of infection risks. Consultation with policymakers and stakeholders responsible for safeguarding public health will be critical in determining the next steps for application of these findings to reducing the risks of vCJD transmission.
Acknowledgments
The authors thank the staff at the Institute for Animal Health, Compton, and The Roslin Institute for animal care and technical assistance. The authors also thank Barry Bradford (The Roslin Institute, The University of Edinburgh) for preparing, unblinding, and decoding a mini panel of samples from the bovine spongiform encephalopathy sheep archive.
The results presented in this paper are based on independent research commissioned and funded by the Policy Research Programme of the Department of Health and Social Care (https://www.nihr.ac.uk/explore-nihr/funding-programmes/policy-research.htm). The award (“Comparative evaluation of the performance of proposed diagnostic tests for vCJD in preclinical blood samples”; National Institute for Health and Care Research [NIHR] reference: PR-R17-0916-23006) was made to E.F.H.
The views expressed in the publication are those of the authors and not necessarily those of the Department of Health and Social Care, “arm’s-length” bodies, or other government departments. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Authorship
Contribution: E.F.H. designed the research and obtained funding; C.M.T., F.A., M.K.F.S., and O.A. performed experiments; C.M.T., D.B., E.F.H., M.E.A., M.K.F.S., and O.A. analyzed data and results; O.A. was responsible for mouse bioassays; M.E.A. analyzed bioassay data; K.L. and J.K.C. were responsible for assembling, blinding, and decoding panels of blood samples; C.M.T. and E.F.H. prepared figures and wrote the manuscript; and all other authors contributed to editing the final draft.
Conflict-of-interest disclosure: Établissement Français du Sang holds patents for “Nanobeads covered with plasminogen as a direct support for cyclic amplification of the prion protein PrPSc” (US 13/425 806, FR 1152298, EP 12160615.6, 2011). All other authors declare no conflict of interest.
Correspondence: E. Fiona Houston, The Roslin Institute, The Royal (Dick) School of Veterinary Studies, The University of Edinburgh, Easter Bush Campus, Edinburgh EH25 9RG, United Kingdom; email: fiona.houston@roslin.ed.ac.uk.
References
Author notes
All data generated or analyzed during this study are included in this published article and its supplemental files, available with the online version of this article. Original data from individual experiments are available from the corresponding author, E. Fiona Houston (fiona.houston@roslin.ed.ac.uk), on reasonable request.
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal