In this issue of Blood, Arrieta-Bolaños et al report on the effects of HLA-DPB1 T-cell epitope (TCE) mismatch, including mismatch directionality, in unrelated hematopoietic cell transplantation (UR-HCT). They demonstrate that noncore permissive graft-versus-host (GVH) mismatches reduce disease relapse risk, whereas core permissive mismatches minimize acute graft-versus-host disease (GVHD) risk.1
In UR-HCT, HLA-A, -B, -C, and -DRB1 (HLA-8/8)-matched or HLA-A, -B, -C, -DRB1, and -DQB1 (HLA-10/10)-matched donors are selected as optimal donors. For HLA-DPB1, retrospective clinical studies have provided clear evidence that patient–donor HLA-DPB1 mismatch is associated with an increased risk of acute GVHD and a reduced risk of leukemia relapse. Recently, 2 different models have been proposed for the selection of HLA-DPB1-mismatched unrelated donors, namely, the HLA-DPB1 TCE and the HLA-DP expression models (see figure panel A). The single-nucleotide polymorphism rs9277534 located in the 3′ untranslated region (3′UTR) of the HLA-DPB1 gene is associated with the expression level of HLA-DP. In the expression model, the risk of acute GVHD induced by patient–donor HLA-DPB1 mismatch is influenced by the rs9277534 variation.2 Evolutionary analysis using sequencing data of the entire HLA-DPB1 region revealed that the HLA-DPB1 gene is highly conserved from exon 3 to the 3′UTR and may provoke acute GVHD in a different manner compared with the TCE model, which reflects exon 2 polymorphisms.3
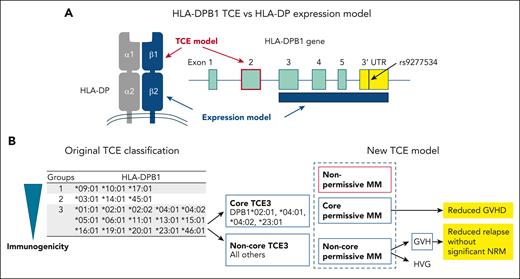
Models for the selection of HLA-DPB1-mismatched unrelated donors. (A) HLA-DPB1 TCE and HLA-DP expression models for the selection of HLA-DPB1-mismatched unrelated donors. (B) Original classification of HLA-DPB1 TCE groups and new HLA-DPB1 TCE expression models proposed by Arrieta-Bolaños et al. NRM, nonrelapse mortality.
Models for the selection of HLA-DPB1-mismatched unrelated donors. (A) HLA-DPB1 TCE and HLA-DP expression models for the selection of HLA-DPB1-mismatched unrelated donors. (B) Original classification of HLA-DPB1 TCE groups and new HLA-DPB1 TCE expression models proposed by Arrieta-Bolaños et al. NRM, nonrelapse mortality.
The HLA-DPB1 TCE model was originally defined in vitro based on the cross-reactivity pattern of a T-cell clone obtained from a patient at the time of rejection after HLA-DPB1-mismatched allogeneic transplantation.4 In the original TCE model, HLA-DPB1 alleles were classified into 3 TCE groups according to the strength of their immunogenicity. HLA-DPB1 alleles in the same TCE group share similar amino acid sequences in the peptide-binding regions of the HLA-DP molecule. If the patient’s and donor’s mismatched HLA-DPB1 alleles belong to the same TCE group, the HLA-DPB1-allele mismatches were defined as “permissive,” whereas if they were in different groups, they were defined as “nonpermissive.” Nonpermissive mismatches were associated with a higher risk of acute GVHD, mortality, or both in UR-HCT.5,6 The original HLA-DPB1 TCE classification algorithm was updated to predict HLA-DPB1 permissive or nonpermissive mismatches between patients and donors, based on the functional distance scores of 10 polymorphic amino acids located within the hypervariable region of the HLA-DPB1 gene.7 In other words, an HLA-DPB1 mismatch with a high functional-distance score is nonpermissive. The National Marrow Donor Program guidelines recommend the selection of an HLA-DPB1-matched donor or an HLA-DPB1 permissive mismatched donor in UR-HCT, in accordance with the updated HLA-DPB1 TCE algorithm.8
Because the TCE model is associated with GVHD and survival, Arrieta-Bolaños et al attempt to further refine the grouping of the TCE classification (see figure panel B). Prior to the study reported in this issue of Blood, the authors had investigated the structural hierarchies of HLA-DPB1 alleles, which were characterized using multidimensional scaling techniques based on polymorphic amino acid positions that encompass all the hypervariable regions in the HLA-DP molecules.9 Among the HLA-DPB1 alleles in the TCE3 group, HLA-DPB1∗02:01, 04:01, 04:02, and 23:01 (core TCE3 alleles) form a distinct cluster separate from the other alleles (noncore TCE3 alleles). Arrieta-Bolaños et al found that TCE3 core alleles elicit lower CD4+ T-cell alloreactive responses compared with TCE3 noncore alleles and that incompatibility between patient and donor TCE3 core alleles (core permissive mismatch) is associated with a reduced risk of acute GVHD and transplant-related mortality compared with a nonpermissive mismatch.
In the study reported in this issue of Blood, the authors incorporated directionality into the classification of HLA-DPB1 TCE mismatches. Compared with HLA-DPB1 allele-matched pairs, noncore permissive mismatch pairs showed significantly higher risk of acute GVHD and significantly reduced risk of relapse, but there were no significant differences in the risk of acute GVHD and relapse between core permissive mismatch and allele-matched pairs. When permissive and nonpermissive mismatch pairs were stratified into GVH and host-versus-graft (HVG) directions, both HVG and GVH nonpermissive mismatches were associated with an increased the risk of acute GVHD, although only nonpermissive GVH mismatches were associated with reduced relapse and increased nonrelapse mortality compared with allele-matched pairs. Among permissive mismatch pairs, noncore permissive GVH mismatches were associated with a lower risk of relapse and a higher risk of acute GVHD compared with allele-matched pairs. Meanwhile, neither core nor noncore mismatches were associated with nonrelapse mortality. The authors concluded that they have shown for the first time that directional noncore permissive GVH drives effective graft-versus-leukemia responses without a significant increase in nonrelapse mortality. This suggests that tailored unrelated donor selection should be a clinical priority for patients with hematological malignancies.
The novelty of this study lies in its incorporation of directionality into the classification of the HLA-DPB1 TCE model. Although further validation of this new model is needed, it may be applicable to unrelated donor selection according to patient risk, including patients with hematologic malignancies in terms of GVL efficacy and those with nonhematologic malignancies in terms of reduced risk of GVHD.
Because it is insufficient to predict the transplant outcomes associated with HLA-DPB1 mismatch using a single mismatch model, a new donor-selection tool that integrates the TCE model with the expression model has recently been reported.10 In the future, it will be necessary to integrate the updated TCE model into all relevant analyses.
Conflict-of-interest disclosure: S.M. declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal