In this issue of Blood, Abdelbaky and colleagues report the development of a combined epigenetic and immunogenetic signature to study individuals with high-count monoclonal B-cell lymphocytosis (HC-MBL).1 Using this signature, they define a new high-risk group with significantly increased likelihood of progression to chronic lymphocytic leukemia (CLL) compared with low-risk individuals.
Twenty-five years ago, 2 seminal articles were published in Blood, fundamentally altering the landscape of CLL research.2,3 These studies demonstrated that patients with CLL with somatically hypermutated immunoglobulin heavy variable (IGHV) genes (M-CLL; 60%-70% of patients) experienced better outcomes than patients with unmutated IGHV genes (U-CLL; 30%-40% of patients). Over time, the IGHV gene mutational status has been established as a robust prognostic and predictive marker. As a result, contemporary guidelines recommend IGHV gene analysis for all patients before initiating treatment.
DNA methylation arrays revealed early on that U-CLL and M-CLL display distinct methylation profiles. In a pivotal study from 2012, Kulis et al compared the methylation signature in CLL with different B-cell differentiation stages.4 This led to the identification of 2 major “epitypes,” one resembling naive B cells, largely corresponding to U-CLL (termed naive B-cell-like CLL), and the other more similar to memory B cells, overlapping with M-CLL (designated memory B-cell-like CLL). An intermediate epitype (i-CLL) with an intermediate methylation pattern and a corresponding intermediate outcome was also identified. In a similar study, based on the level of DNA methylation programming, Oakes et al also identified 3 prognostic subgroups, which were named the high-programmed (HP), intermediate-programmed (IP), and low-programmed (LP) epitypes.5
Further characterization of the intermediate epitype revealed an enrichment of CLL cases with borderline IGHV mutational status (97%-97.9% identity) or SF3B1 mutations or belonging to stereotyped subset 2.6,7 Subsequently, Maity et al identified a critical somatic mutation (G>C) in the IGLV3-21 gene at position R110 in subset 2, crucial for homeotypic B-cell receptor interaction.8 Extended analysis showed that nonsubset 2 cases with the IGLV3-21R110 mutation had a similarly poor prognosis as subset 2. Furthermore, a recent study suggested that intermediate patients can be regrouped depending on IGLV3-21R110 status; patients with IGLV3-21R110-mutated i-CLL demonstrated a poor outcome similar to patients with naive B-cell-like CLL, and patients with IGLV3-21R110-wildtype i-CLL had a similar prognosis as patients with memory B-cell-like CLL.7
Healthy individuals may harbor clonal B-cell expansions of varying sizes, termed monoclonal B-cell lymphocytosis, a precursor condition always preceding CLL.9 The likelihood of MBL increases with age and in patients with first-degree relatives with CLL. MBL can be categorized into low-count MBL (<0.5 × 109 clonal B cells per L) and HC-MBL (>0.5 × 109 to <5 × 109 per L). Although the majority of HC-MBL cases exhibit favorable-prognostic features (IGHV-mutated and low-risk genomics), an estimated 1%-2% of individuals with HC-MBL will develop CLL requiring treatment.1,9 Considering the relatively low proportion of HC-MBL cases developing high-risk disease, identifying new biomarkers is crucial for early-stage identification of those at high risk of progression.
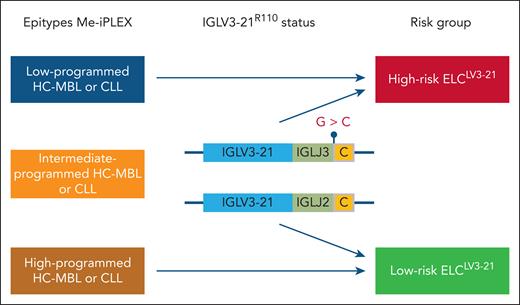
In the study by Abdelbaky et al, information from DNA methylation profiling was combined with the assessment of the IGLV3-21R110 status to create a joint epigenetic and immunogenetic signature termed the epigenetic and light chain IGLV3-21 immunoglobulin (ELCLV3-21) signature (see figure). Using their previously developed methylation-iPLEX (Me-iPLEX) assay,6 which interrogates the methylation status of 7 loci, the majority of HC-MBL (65%) belonged to the good-prognostic HP epitype, and the IP epitype and LP epitypes were seen in 19% and 16% of individuals, respectively. Sequencing of the λ light-chain rearrangements revealed that 19% of individuals in the IP epitype carried the IGLV3-21R110 mutation. Notably, individuals with the IP epitype and the IGLV3-21R110 mutation had a similar time to CLL progression and time to first treatment (TTFT) as those with the LP epitype, leading to the grouping of these cases into an ELCLV3-21 high-risk group. In multivariable analysis, including ELCLV3-21 and IGHV mutation status, only the ELCLV3-21 signature remained significant. Similarly, when including CLL-international prognostic index (IPI) and ELCLV3-21, only ELCLV3-21 retained its status as an independent risk factor.
The combined epigenetic and immunogenetic ELCLV3-21 signature. Using the Me-iPLEX methylation assay, HC-MBL and CLL were divided into LP, IP, and HP epitypes. The IGVL3-21R110 status was assessed by sequencing in IP MBL/CLL. IP cases carrying the IGVL3-21R110 mutation were grouped with the LP epitype into the ELCLV3-21 high-risk group, and IGVL3-21R110-wild-type IP cases were merged with the HP epitype into the ELCLV3-21 low-risk group.
The combined epigenetic and immunogenetic ELCLV3-21 signature. Using the Me-iPLEX methylation assay, HC-MBL and CLL were divided into LP, IP, and HP epitypes. The IGVL3-21R110 status was assessed by sequencing in IP MBL/CLL. IP cases carrying the IGVL3-21R110 mutation were grouped with the LP epitype into the ELCLV3-21 high-risk group, and IGVL3-21R110-wild-type IP cases were merged with the HP epitype into the ELCLV3-21 low-risk group.
Applying the ELCLV3-21 model to an independent CLL cohort, the ELCLV3-21 high-risk signature was again associated with inferior TTFT and overall survival (OS). In multivariable analysis, including IGHV mutation status, genomic aberrations, and CLL-IPI, ELCLV3-21 maintained significance for TTFT, whereas the IGHV status no longer held significance. Importantly, patients with ELCLV3-21 high-risk HC-MBL had shorter TTFT and OS than patients at low risk of developing CLL, more akin to patients at high risk of developing CLL.
In summary, the authors have identified a new signature to identify individuals with HC-MBL with a particularly high risk of progression to CLL, who also appear to have a shorter OS. Although this new prognostic signature is promising, further validation in independent data sets is crucial, especially considering the relatively low number of ELCLV3-21 high-risk IP individuals included. An in-depth analysis comparing the benefit of the ELCLV3-21 grouping versus only performing IGHV/IGLV gene sequencing is also critical. Moreover, given the diverse treatment regimens received by patients in the collected cohorts over the long time span of the study, the OS data may not be reflective of current treatment practice and might therefore be a less definite finding. Therefore, assessing the impact of the ELCLV3-21 signature in more homogeneously treated cohorts, in particular in relation to targeted therapies such as Bruton tyrosine kinase and B-cell lymphoma 2 inhibitors, is essential.
From a methodological perspective, the authors assert that the Me-iPLEX is a rapid and cost-effective test. However, this assay is not widely used and would need standardization to be implemented in routine diagnostics. Given that other epityping assays include different loci, collaborative efforts among key stakeholders to define the relevant epitypes and nomenclature would be highly beneficial. As the authors point out, their approach can be adapted to next-generation sequencing-based assays, which would be an important development. In fact, using capture-based panel sequencing, it is possible to include epigenetic marks, along with assessment of oncogenic mutations. Although IGHV gene sequencing is performed in routine diagnostics, primarily using Sanger sequencing, the assessment of the IGLV3-21R110 mutation status is currently not part of routine practice. Nevertheless, it would be relatively straightforward to use targeted assays to detect the IGLV3-21R110 mutation or to incorporate this feature in a gene panel design.10 Alternative approaches, such as long-read sequencing, enabling simultaneous analysis of genomic aberrations, and DNA methylation profiles, could also be considered.
Once validated, the ELCLV3-21 signature would be expected to become a highly relevant and useful tool for identifying individuals with a very low risk of CLL progression, as well as high-risk individuals with MBL requiring closer monitoring.
Conflict-of-interest disclosure: R.R. has received honoraria from Abbvie, AstraZeneca, Janssen, Illumina, and Roche.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal