Background :
Venetoclax resistance is a major obstacle to successful treatment of acute myeloid leukemia (AML) and myelodysplastic syndromes (MDS). Its underlying mechanisms are multifaceted and have been linked to monocytic differentiation, reprograming mitochondrial metabolism, and increased expression of BRD4 and other BCL2-like proteins, etc. Our previous studies demonstrated that specific RNA cytosine methyltransferase, namely NSUN1 (aka NOL1, NPO2) and NSUN2, mediate the formation of drug-resistant active chromatin structures (ACS) at nascent RNAs to regulate drug resistance in leukemia cells (Cheng, J.X. et al. Nat Commun, 2018) ( Figure 1A). The Cancer Genome Atlas Program (TCGA) data demonstrated that elevated expression of NSUN1 and NSUN2 is correlated with significantly shorter patients' survival in almost all cancer subtypes. However, due to the lack of the crystal structures of NSUN1 and NSUN2 proteins, no NSUN1/2-targeting therapeutics have been developed yet.
Methods:
AI/supercomputing design of NSUN1/2-targeting drug libraries and analysis of structure activity relationship (SAR).
Newly developed functional technologies, including NSUN1/2-targeting fluorescence releasing assay (FRA), 5-ethynyl uridine click chemistry (EC) and proximity ligation rolling cycle amplification (PL-RCA)-coupled confocal microscopy (CM), flow cytometry (FCM) and RNA sequencing (RNA-seq).
NSUN1/2-targeting functional drug screening (NSUN1/2-FDS) assay and drug growth inhibition (MTT) assay.
H&E histology and immunohistochemistry (IHC).
Enforced NSUN1/2 expression and knockout (KO) cell lines and syngeneic AML mouse models.
Results:
Identification of several ligan-binding modules/surfaces in NSUN1 and NSUN2 proteins and generation of small-molecule libraries to targe these ligand-binding surfaces/modules with AI/supercomputing structural prediction and ligand-receptor interaction simulation.
Identification of several effective small-molecule inhibitors of NSUN1 and NSUN2 by functional screening of the AI-designed drug libraries in venetoclax-resistant human monocytic leukemia (SC), erythroid leukemia (K562) and lung cancer (H82) cells.
Defining the binding modules of the identified NSUN1/2 inhibitors and selecting lead inhibitors through supercomputer/AI-directed SAR analysis.
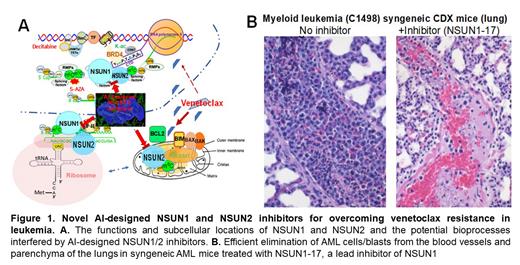
Elucidation of the action mechanisms of NSUN1/2 lead inhibitors thorough disruption of NSUN1/2-mediated ACS in the nucleus and translational complexes (TLC) in the ribosomes and mitochondria ( Figure 1A).
In vivo validation of the efficacies of the identified lead inhibitors using a syngeneic AML mouse model. As shown in Figure 1B, NSUN1-17, one of the lead inhibitors, efficiently eliminated the AML cells/blasts from the blood vessels and parenchyma of the lungs of the syngeneic AML mice.
In addition, our siRNA knock-down experiments demonstrated that downregulation of NSUN1/2 expression re-sensitizes venetoclax-resistant monocytic leukemia cells to a low dose of venetoclax. Our NIH/3T3 soft-agarose assay confirmed the oncogenic transformation capability of NSUN1 and NSUN2. We have also successfully established the homozygous ( Nsun2-/-) mouse model with markedly reduced body weight, but a normal lifespan, compared to the heterozygous mice ( Nsun2+/-).
Conclusion:
By leveraging the Argonne AI/supercomputer and our functional assays, we identified several small-molecule inhibitors that can disrupt the NSUN1/2-mediated ACS and TLC to overcome venetoclax resistance in AML cell lines and syngeneic mice. This study has not only illustrated the importance of NSUN1 and NSUN2 in maintaining oncogenic transcription and translation as well as venetoclax resistance in AML, but also paved the way to the development of novel RNA epigenetics-driven therapeutics for leukemia and other cancers.
Disclosures
No relevant conflicts of interest to declare.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal