Purpose: Myelodysplastic syndromes (MDS), is characterized by clinical heterogeneity and varying prognoses. Reliable prognostic models are essential to stratify MDS patients and optimize clinical management. Although incorporating molecular information into MDS prognostic models improves their predictive ability, limitations of next-generation sequencing panels and potential heterogeneity among MDS patients with different karyotypes hinder their clinical translation.
Methods: In total, 237 normal karyotype (NK) MDS patients from the First Affiliated Hospital of Zhejiang University School of Medicine were used as the derivation set to investigate the molecular frequency and prognostic spectrum. A parsimonious molecular-clinical model was developed with overall survival (OS) as the endpoint and compared with other models using the concordance index as the primary evaluation index. An external independent validation set from the International Working Group for Prognosis in MDS cohort(691 NK MDS patients) was used to evaluate the model generalization ability.
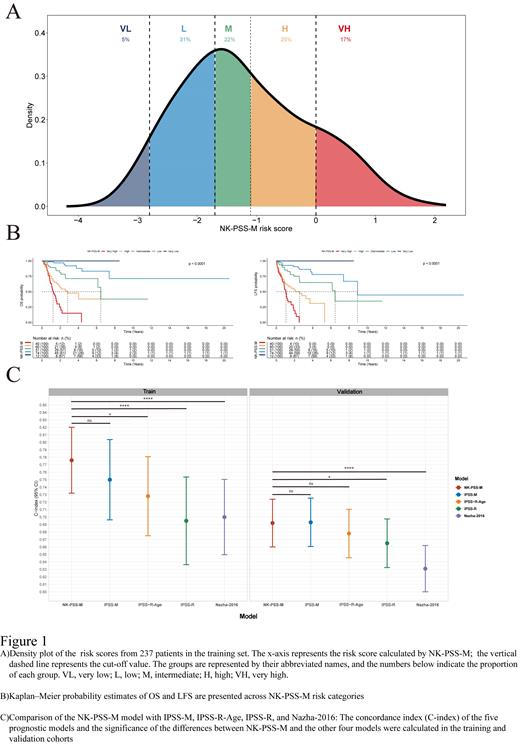
Results: In NK MDS, the gene mutation frequency spectrum is similar to the overall MDS population, the differences mainly lie in the prognostic spectrum. Genes such as TP53, previously associated with MDS prognoses have lost their prognostic value. Therefore, a seven-parameter molecular-clinical prognostic model called the Molecular Prognostic Scoring System for NK MDS (NK-PSS-M) was developed based on a specific molecular prognostic spectrum of NK MDS. The model includes age, hemoglobin, platelet count, blast percentage, CEBPA, RUNX1, and U2AF1. The NK-PSS-M model allows for personalized risk assessment based on patient characteristics, with higher scores indicating a greater risk and poorer prognosis. In addition, we developed a five-class classification system based on these scores, which differentiates patients across risk categories and predicts outcomes, such as OS and LFS(Figure 1 A and B). The performance of NK-PSS-M was comparable to that of the Molecular International Prognostic Scoring System(IPSS-M), and it significantly outperformed IPSS-R and other molecular prediction models based on it. The above results were confirmed in the external independent validation cohort(Figure 1C). Our transcriptomics analysis revealed that the low-risk group defined by NK-PSS-M demonstrated significant activation of the inflammatory pathway, and immune activation, which is consistent with the current understanding of the immunological landscape of MDS.
Conclusion: The NK-PSS-M model improved the risk stratification of non-molecular models and provided a reliable alternative to the IPSS-M. In addition, we demonstrated the molecular heterogeneity of MDS across different cytogenetics and emphasized the importance of developing cytogenetics-specific prognostic models.
Disclosures
No relevant conflicts of interest to declare.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal