In this issue of Blood, Ang et al1 show that human CD20 messenger RNA (mRNA) undergoes alternative splicing to generate distinct 5' untranslated region (5'-UTR) variants, which determine the cell-surface CD20 levels in malignant B cells and the interpatient variability in the expression of this therapeutic target. These variants can also be used as an escape mechanism from anti-CD20 therapies.
The approval of rituximab, the anti-CD20 antibody, by the Food and Drug Administration in 1997 was a conceptual breakthrough for the treatment of “mature” B-cell malignancies. Based on rituximab's success, newly engineered anti-CD20 monoclonal antibodies have been manufactured, followed by the development of anti-CD20 chimeric antigen receptor T cells and anti-CD20/CD3 bispecific T cell engagers (such as mosunetuzumab). Although CD20 is an ideal therapeutic target, it is still unclear how its expression is regulated in normal or in malignant B cells. The need to understand CD20 regulation is underscored by the surprising observation that B-cell receptor inhibitors greatly reduce CD20 levels by interfering with its transcriptional regulation.2,3 Rituximab efficacy is highly dependent on cell-surface CD20 levels,4,5 and the reduction of CD20 levels at least partially explains the resistance to anti-CD20 antibodies and the lack of clinical benefit of the addition of rituximab to the Bruton tyrosine kinase inhibitor ibrutinib in chronic lymphocytic leukemia (CLL).6
Several different MS4A1 mRNA transcripts (encoding CD20) were identified when the structure of MS4A1 was characterized in 1989.7 Tedder et al7 noted that the dominant MS4A1 mRNA variant is 2.8 kilobase long and uses all 8 exons encoded by MS4A1, whereas a second variant is 263 nucleotides shorter with the exon 1 spliced into an internal 3' splice site within exon 3, thereby skipping exon 2. However, all identified transcripts were shown to be translated into identical full-length CD20 proteins as the translation start codon is localized within exon 3, and the importance of these splicing variants remained unclear. Moreover, other MS4A1 alternative splice variants that involve the open reading frame (exon 3-8) were identified in malignant or Epstein-Barr virus (EBV) transformed B cells, some of them encoding truncated CD20 protein, resulting in impaired efficacy of anti-CD20 antibodies.8
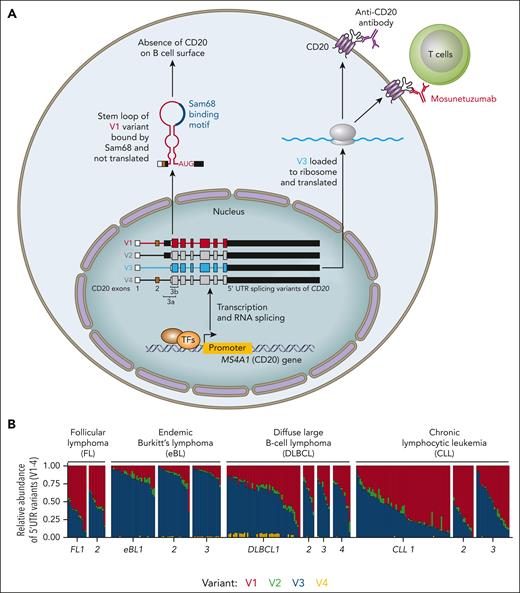
To identify MS4A1 mRNA isoforms, Ang et al performed Nanopore long-read sequencing on Raji cell lines and found three 5'-UTR isoforms corresponding to previously identified transcripts (V1, V2, and V3) and also a 4th rarer undescribed 5'-UTR variant (V4) (see figure panel A). Similar to the observations by Tedder et al,7 all of these mRNA isoforms had distinct 5'-UTRs with a splicing event occuring between exon 1 and exon 3 (or internal splice site in exon 3) but had identical coding sequences. The authors used publicly available short-read RNA sequencing data from various B-cell malignancies (B-cell acute lymphoblastic leukemia, endemic Burkitt lymphoma [BL], CLL, diffuse large B-cell lymphoma [DLBCL], and follicular lymphoma [FL]) and performed the analysis of exon-exon junctions for the V1-4 variants. This revealed that V1 and V3 together represent >90% of MS4A1 isoforms and the ratio of V3 to V1 has a wide distribution within samples with the same diagnosis. Among the B-cell neoplasms, EBV+ endemic BL stood out as having a relatively higher V3:V1 ratio, and this V1-to-V3 shift was recapitulated when normal B cells were infected by EBV in vitro. The change in isoforms toward V3 (or V4) after EBV infection cooccurred with elevation in cell-surface CD20 levels, and this is especially notable since EBV infection downmodulated the total MS4A1 mRNA levels. This led the authors to hypothesize that although the V1 and V3 MS4A1 mRNA isoforms have identical coding sequences, they possess distinct 5'-UTRs that can alter mRNA turnover and/or translation, a mechanism previously noted by the same laboratory for CD22.9 Indeed, relative to V3 and V4, a smaller fraction of V1 and V2 molecules was associated with polysomes reflecting decreased propensity for translation. Overexpression of engineered vectors with all variants (V1-4) demonstrated that inefficient V1 and V2 transcript translation is rate-limiting for CD20 protein production. In some of these experiments, the overexpression of isolated V1 isoform in a cell line did not lead to any detectable CD20 protein levels, whereas V3 isoform led to a massive increase in CD20 protein, suggesting a major role for the V3:V1 ratios in determining cell surface CD20 levels. Inefficient V1 (and V2) translation implied the presence of repressive 5'UTR elements. Detailed analysis revealed that the exon 3a sequence contained in V1 and V2 isoforms harbors multiple upstream open reading frames (uORFs) and that the exon 3a-b boundary immediately upstream of the MS4A1 start codon also forms a stem-loop secondary structure (see figure panel A). Deletion mutants and engineered variants with mutated start codons in uORFs showed that these elements are responsible for the negligible CD20 protein produced from V1 and V2 mRNAs. The exon 3a loop structure contains binding sites for Sam68, a splicing factor known to bind to AU-rich sequences. Sam68 knockdown in Raji cells resulted in the redistribution of 5'-UTR variants, suggesting that Sam68 is at least partly responsible for selecting MS4A1 isoforms.
The 5'-UTR splicing variants of MS4A1 (encoding CD20) and their frequency in mature B-cell malignancies. (A) The transcription of MS4A1 is regulated by multiple cell signals and transcription factors (TFs). Four 5'-UTR isoforms (V1-V4) of MS4A1 have been identified resulting from distinct splicing events between exon 1 and 3 (internal splice site), but all have identical coding sequences (translation start codon is localized within exon 3). The V3 and V4 represent translation proficient variants, whereas inefficient translation of V1 and V2 transcripts is rate-limiting for CD20 protein production. The V1 and V2 variants harbor multiple upstream open reading frames and a stem-loop secondary structure containing binding sites for Sam68 protein. (B) The frequency of V1-4 variants of MS4A1 in samples from multiple cohorts of B-cell malignancies (FL, n = 2; endemic BL, n = 3; DLBCL, n = 4; CLL, n = 3). The V1 and V3 represent together >90% of MS4A1 isoforms, and their ratio has a wide distribution within samples from the same diagnosis. Professional illustration by Patrick Lane, ScEYEnce Studios.
The 5'-UTR splicing variants of MS4A1 (encoding CD20) and their frequency in mature B-cell malignancies. (A) The transcription of MS4A1 is regulated by multiple cell signals and transcription factors (TFs). Four 5'-UTR isoforms (V1-V4) of MS4A1 have been identified resulting from distinct splicing events between exon 1 and 3 (internal splice site), but all have identical coding sequences (translation start codon is localized within exon 3). The V3 and V4 represent translation proficient variants, whereas inefficient translation of V1 and V2 transcripts is rate-limiting for CD20 protein production. The V1 and V2 variants harbor multiple upstream open reading frames and a stem-loop secondary structure containing binding sites for Sam68 protein. (B) The frequency of V1-4 variants of MS4A1 in samples from multiple cohorts of B-cell malignancies (FL, n = 2; endemic BL, n = 3; DLBCL, n = 4; CLL, n = 3). The V1 and V3 represent together >90% of MS4A1 isoforms, and their ratio has a wide distribution within samples from the same diagnosis. Professional illustration by Patrick Lane, ScEYEnce Studios.
Altogether, more than 30 years after the work by Tedder et al,7 this study deciphered the effect of MS4A1 mRNA isoforms on this mRNA's translation efficacy. The evolutionary importance of this regulatory mechanism remains unknown. Likewise, it is not known if this mechanism also applies to any of the other 18 members of this gene family. Practically, the findings reported here have direct implications for understanding why some B-cell malignancies or individual patients have lower pre-therapy CD20 levels. Indeed, screening V1-4 isoforms in large cohorts of patients' samples revealed that some have a near absence of translation-proficient V3 variant (see figure panel B). The CD20 levels are heterogeneous within the intraclonal cell subpopulations in an individual patient,10 and it remains to be determined whether V3:V1 ratios change during B-cell exposure to signals known to regulate CD20 levels on malignant B cells, such as interleukin-4 or SDF1 chemokine.2,3 Moreover, changes in V3:V1 might be a potential escape mechanism for anti-CD20 therapies. Indeed, Ang et al observed a V3-to-V1 shift in 2 out of 4 patients with mosunetuzumab-resistant follicular lymphoma.
Finally, the authors used morpholino antisense oligonucleotides to enhance CD20 translation by modulating alternative splicing of noncoding exons of MS4A1. Such treatment induced shifts in MS4A1 splicing, namely increasing V3 and V4 variants and elevated CD20 protein. This resulted in a >10-fold decrease in rituximab inhibitory concentration in 2 out of 3 tested cell lines, providing the first steps toward RNA-based therapeutics.
Conflict-of-interest disclosure: The author declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal