Key Points
SLFN14 K219N platelets and MKs exhibit rRNA degradation with compensation for ribosome biogenesis via the mTORC1 pathway.
SLFN14-related TP results from defective megakaryopoiesis with abnormal mitochondria in MKs.
Abstract
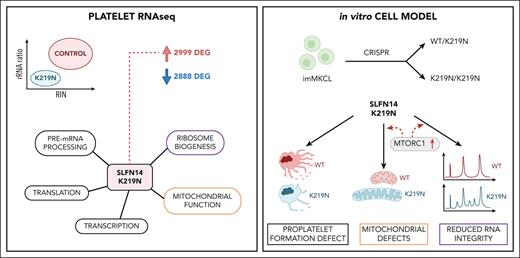
Pathogenic missense variants in SLFN14, which encode an RNA endoribonuclease protein that regulates ribosomal RNA (rRNA) degradation, are known to cause inherited thrombocytopenia (TP) with impaired platelet aggregation and adenosine triphosphate secretion. Despite mild laboratory defects, the patients displayed an obvious bleeding phenotype. However, the function of SLFN14 in megakaryocyte (MK) and platelet biology remains unknown. This study aimed to model the disease in an immortalized MK cell line (imMKCL) and to characterize the platelet transcriptome in patients with the SLFN14 K219N variant. MK derived from heterozygous and homozygous SLFN14 K219N imMKCL and stem cells of blood from patients mainly presented with a defect in proplatelet formation and mitochondrial organization. SLFN14-defective platelets and mature MK showed signs of rRNA degradation; however, this was absent in undifferentiated imMKCL cells and granulocytes. Total platelet RNA was sequenced in 2 patients and 19 healthy controls. Differential gene expression analysis yielded 2999 and 2888 significantly (|log2 fold change| >1, false discovery rate <0.05) up- and downregulated genes, respectively. Remarkably, these downregulated genes were not enriched in any biological pathway, whereas upregulated genes were enriched in pathways involved in (mitochondrial) translation and transcription, with a significant upregulation of 134 ribosomal protein genes (RPGs). The upregulation of mitochondrial RPGs through increased mammalian target of rapamycin complex 1 (mTORC1) signaling in SLFN14 K219N MK seems to be a compensatory response to rRNA degradation. mTORC1 inhibition with rapamycin resulted in further enhanced rRNA degradation in SLFN14 K219N MK. Taken together, our study indicates dysregulation of mTORC1 coordinated ribosomal biogenesis is the disease mechanism for SLFN14-related TP.
Introduction
SLFN14 is an endoribonuclease protein belonging to the Schlafen protein family. Members of this family have been shown to regulate cell proliferation, differentiation and migration, virus replication, and interferon signaling.1 In humans, 5 SLFN proteins have been identified: SLFN5, SLFN11, SLFN12, SLFN13, and SLFN14.1 Disease-causing missense variants in SLFN14 were identified in the UK Genotyping and Phenotyping of Platelets study in 12 patients from 3 unrelated families with thrombocytopenia (TP).2 Obvious bleeding tendencies were reported in all patients whereas laboratory testing showed mild defects in platelet aggregation and adenosine triphosphate (ATP) secretion, and platelet counts that were mildly reduced. The identified variants affect 3 consecutive amino acids that are part of the “ATPase associated with diverse cellular activities” protein domain: K128E, K219N, and V220D.2,3 In a subsequent study by Marconi et al, R223W was discovered in a family with macrothrombocytopenia.4 In this case report, in vitro megakaryopoiesis was studied using blood-derived stem cells from 2 related patients. Megakaryocyte (MK) cultures from patients produce less proplatelets with shorter elongation and reduced ramification of their shafts.4 SLFN14 protein expression studies in platelets from patients and transfected cells showed that these heterozygous missense variants caused a >50% decreased expression, indicating a possible dominant negative effect on wild-type (WT) SLFN14.2,4,5 A CRISPR knock-in mouse model of SLFN14 K208N (homologous to K219N) was created.6 Remarkably, heterozygous K208N mice present with microcytic erythrocytosis, hemolytic anemia, splenomegaly, abnormal thrombus formation, and a mildly increased platelet size although platelet count, function, and morphology were unaffected. This study suggested that SLFN14 is a species-specific regulator of platelets and erythrocytes. In 2015, Pisareva et al showed that, in rabbit reticulocytes, SLFN14 acts as an endoribonuclease that cleaves RNA, with a preference for ribosomal RNA (rRNA) as well as ribosome-associated messenger RNA (mRNA) molecules.7 The endoribonuclease activity of SLFN14 was confirmed by Fletcher et al, who showed that SLFN14 binds to ribosomes in DAMI and HEK293T cells overexpressing either WT or mutant SLFN14.5 However, in these cellular assays, none of the investigated mutants (K218E, K219N, and V220D) showed a functional effect on ribosomal binding or rRNA degradation. The mutant SLFN14 proteins showed partial misfolding, likely resulting in posttranslational degradation. The authors hypothesized that mutant SLFN14 forms hetero-oligomers with the WT protein, leading to the degradation of both WT and mutant proteins, thus causing a dominant negative effect.2,4,5 These cellular studies were performed using SLFN14 overexpression studies that might have influenced the normal function of this protein. DAMI cells are also megakaryoblastic leukemia cells that are biologically different from primary MK.8 The role of mutant SLFN14 in megakaryopoiesis and platelet dysfunction remains unknown.
We developed heterozygous and homozygous K219N SLFN14 immortalized MK cell lines (imMKCLs) using CRISPR/Cas9 to study the effect of this variant on platelet formation. These imMKCL cells have previously been developed for platelet transfusion purposes.9 In addition, the platelet transcriptome was studied in 2 patients with TP with the K219N SLFN14 variant using platelet RNA sequencing (RNA-seq). Molecular and morphological studies were performed using the imMKCL models to study the defective pathways identified in RNA-seq data.
Materials and methods
Platelet functional testing and MK studies
WES and platelet RNA-seq
Patients IV.1 and IV.2 were enrolled in the National Institute for Health Research BioResource Rare Diseases study for whole-exome sequencing (WES).12 Platelets of 2 patients and 19 different healthy individuals (1 donor was sampled at 2 time points) were used for RNA extraction as described.13 Total RNA-seq was performed by Macrogen Europe. Approval was obtained from the UZLeuven Ethical committee for WES (ML3580/S50025) and RNA-seq (S63666) and informed consent was obtained from all individuals. RNA-seq analyses have been described in the supplemental Methods, available on the Blood website.
Genetic modification and differentiation of the imMKCL
The SLFN14 c.657A>T variant was introduced in the imMKCL using CRISPR/Cas9 to generate 2 homozygous and 2 heterozygous SLFN14 K219N cell lines and RNA interference–mediated SLFN14-depleted imMKCL cells were generated as described in the supplemental Methods.
Flow cytometry
Platelets, primary MK, and imMKCL cells were analyzed on a BD FACS Canto cytometer using the FCS Express software (De Novo Software). Analysis of propidium iodide–stained cells was performed as described.14 All antibodies are listed in the supplemental Methods.
Imaging
Cells attached to coverslips were stained as described in supplemental Methods. An LSM 700 (Zeiss) confocal microscope was used to image Mitotracker Red and ATP5E staining. Brightfield, SiR-tubulin, and SiR-DNA imaging of live cells was performed using the Cytation 5 Cell Imaging Multi-Mode Reader (Biotek). Electron microscopy (EM) samples were prepared as described in supplemental Methods and imaged using a JEM 1400-LaB6 Electron Microscope (JEOL Ltd Japan). Images were analyzed using the ImageJ software.
Immunoblot analysis
Patient platelet samples or imMKCL cells were lysed to obtain protein samples as described.14 Blots were imaged on a ChemiDoc XRS+ imager and quantified using Image Lab software (BioRad).
Quantification of proplatelet formation
Proplatelet-forming MK was counted on day 5 by a blinded assessment. MK was imaged randomly in uncoated 96-well plates using a Cytation 5 Cell Imager. Nine images were stitched per well and analyzed in triplicate for 3 independent differentiation experiments.
Seahorse assay
The mitochondrial oxygen consumption rate (OCR) was measured in day 4 MK using a Seahorse FX HS Mini Analyzer, as described in the supplemental Methods.
RNA quantification using the Agilent 2100 BioAnalyzer
RNA was extracted from the cells using Trizol. RNA quality was determined using an Agilent RNA 6000 Nano Kit on a 2100 Bioanalyzer system as described by the manufacturer.
Rapamycin mTOR inhibitor assays
Rapamycin (CAS 53123-88-9, Calbiochem) was added as a single dose to imMKCL cells (100 nM) 24 hours after the start of differentiation.
Results
Clinical phenotype and platelet and MK characteristics of SLFN14-deficient patients
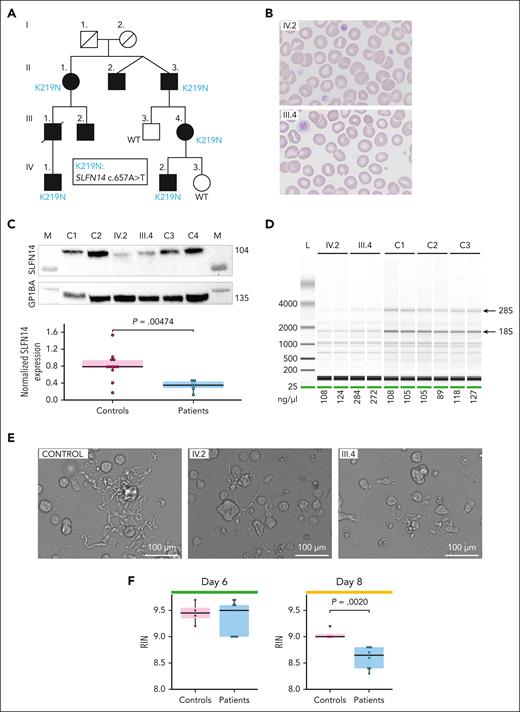
Patients IV.1 and IV.2, from a pedigree followed for an extensive history of bleeding symptoms, which was disproportionate to the degree of TP (Figure 1A), were enrolled in the National Institute for Health Research BioResource Rare Disease study to determine the genetic cause of the bleeding phenotype using WES. Platelet testing showed reduced aggregation responses to adenosine diphosphate and collagen, and mildly reduced ATP secretion (Table 1). The expression of CD62P (α-granule marker) and CD63 (dense granule marker) was reduced mainly after activation with collagen (Table 1). Patients also showed a trend toward elevated erythrocyte counts. Red blood cells were smaller and presented with an obvious increase in red cell distribution width (Table 1), in line with microcytic erythrocytosis found in SLFN14-K208N knock-in mice.6 WES identified the presence of the pathogenic heterozygous SLFN14 variant c.657A>T, resulting in K219N (Figure 1A).2 Sanger sequencing confirmed the presence of the variant in 3 other affected family members (supplemental Figure 1). Flow cytometry analysis showed a mild decrease in CD41+ (integrin ɑIIb), but not CD42a+ (glycoprotein IX), platelets from patients IV.1, IV.2, and III.4 compared with control platelets (supplemental Figure 2). Blood smears showed the presence of some larger platelets in patients IV.2 (21%) and III.4 (9.5%) (Figure 1B). Flow cytometric analysis of CD42b+ platelets in whole blood showed no major differences in overall size (supplemental Figure 3). EM analysis revealed the presence of segregosomes in a few platelets, membrane inclusions, and small dense granules with an empty border (supplemental Figure 4). SLFN14 protein expression was measured by immunoblot analysis using platelet lysates from controls, patients IV.2 and III.4 (Figure 1C), and unaffected relative IV.3 (supplemental Figure 5). A nearly 50% decrease in SLFN14 expression was detected in the presence of the K219N variant. Platelet lysates showed variable expression levels of α- and dense granule markers (supplemental Figure 6). The quantification of platelet RNA using the Bioanalyzer showed the presence of 28S and 18S rRNA degradation fragments in samples from both patients (Figure 1D; supplemental Figure 7).
Pedigree and platelet characteristics of SLFN14 K219N carriers. (A) Pedigree showing affected family members (black symbols) with TP. The presence of the K219N SLFN14 variant is indicated when tested. (B) Peripheral blood smears (May-Grünwald-Giemsa stain; original magnification ×500) of patients IV.2 and III.4 showing platelet anisocytosis with the presence of small and large, hypogranular, platelets. Note the presence of microcytic erythrocytes. (C) Representative immunoblot analysis of SLFN14 protein expression in platelets from patients IV.2 and III.4 and 2 unrelated age- and gender-matched controls for each patient (top). The expression of GP1BA was used as a loading control and for normalization. Marker lanes are indicated as M. Quantification of triplicate immunoblots show a significant reduction in SLFN14 expression in platelets from patients with SLFN14 compared with 4 healthy controls (bottom). Boxplots represent the middle 50% of the data points, with 25% outliers as whiskers and the mean as a black horizontal line. All individual data points were added as dots to the graph. Three technical replicates were performed for each sample. P values were determined using the Wilcoxon test. (D) Representative electrophoresis image showing rRNA degradation of the 28S and 18S peaks (arrows) for platelet RNA samples of patients IV.2 and III.4, compared with platelet RNA from 3 unrelated controls. The concentration of total RNA (ng/μL) for the different samples was added below the figure. (E) Representative brightfield microscopy images illustrating proplatelet formation in blood-derived stem cell cultures after 8 days of MK differentiation showing shorter proplatelets with reduced ramification in cultures from patients IV.2 and III.4, compared with the control. Scale bar, 100 μm. (F) Quantification of the RIN values obtained for RNA samples from days 6 and 8 MK differentiated from blood-derived stem cells of patients IV.2 and III.4 vs a control (4 technical replicates for control days 6 and 8, patient IV.2 days 6 and 8, and patient III.4 day 8, 5 technical replicates for patient III.4 day 6). Statistical analyses were performed using the Wilcoxon test.
Pedigree and platelet characteristics of SLFN14 K219N carriers. (A) Pedigree showing affected family members (black symbols) with TP. The presence of the K219N SLFN14 variant is indicated when tested. (B) Peripheral blood smears (May-Grünwald-Giemsa stain; original magnification ×500) of patients IV.2 and III.4 showing platelet anisocytosis with the presence of small and large, hypogranular, platelets. Note the presence of microcytic erythrocytes. (C) Representative immunoblot analysis of SLFN14 protein expression in platelets from patients IV.2 and III.4 and 2 unrelated age- and gender-matched controls for each patient (top). The expression of GP1BA was used as a loading control and for normalization. Marker lanes are indicated as M. Quantification of triplicate immunoblots show a significant reduction in SLFN14 expression in platelets from patients with SLFN14 compared with 4 healthy controls (bottom). Boxplots represent the middle 50% of the data points, with 25% outliers as whiskers and the mean as a black horizontal line. All individual data points were added as dots to the graph. Three technical replicates were performed for each sample. P values were determined using the Wilcoxon test. (D) Representative electrophoresis image showing rRNA degradation of the 28S and 18S peaks (arrows) for platelet RNA samples of patients IV.2 and III.4, compared with platelet RNA from 3 unrelated controls. The concentration of total RNA (ng/μL) for the different samples was added below the figure. (E) Representative brightfield microscopy images illustrating proplatelet formation in blood-derived stem cell cultures after 8 days of MK differentiation showing shorter proplatelets with reduced ramification in cultures from patients IV.2 and III.4, compared with the control. Scale bar, 100 μm. (F) Quantification of the RIN values obtained for RNA samples from days 6 and 8 MK differentiated from blood-derived stem cells of patients IV.2 and III.4 vs a control (4 technical replicates for control days 6 and 8, patient IV.2 days 6 and 8, and patient III.4 day 8, 5 technical replicates for patient III.4 day 6). Statistical analyses were performed using the Wilcoxon test.
Clinical and hematological characteristics
| Identity . | Bleeding symptoms . | Erythrocyte count (normal: 4 × 106/μL to 5.2 × 106/μL) . | Red cell distribution width (normal: 11.7% -14.5%) . | Mean corpus volume (normal: 77-95 fL) . | Platelet count (normal: 150 × 109/L to 450 × 109/L) . | Mean platelet volume (normal: 8-12 fL) . | Immature platelet fraction (normal: 1.1% -6.1%) . | Platelet aggregation defect (amplitude <50%) for the indicated agonist . | ATP secretion (normal: 1.2-2.8 μM) . | Flow cytometry for CD62P with MFI basal (normal: 97-196) and after activation with 1 μg/mL convulxin (normal: 305-1606) and 50 μg/mL collagen (normal: 1254-3169) . | Flow cytometry for CD63 with MFI basal (normal: 45-86) and after activation with 1 μg/mL convulxin (normal: 73-121) and 50 μg/mL collagen (normal: 358-507) . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| II.1 | Mild mucocutaneous bleeding symptoms | 4.4 | 17.7 | 88 | 53 | NR∗ | NA | NA | NA | NA | NA |
| II.3 | Mild mucocutaneous bleeding symptoms | 4.5 | 18.5 | 87 | 49 | NR∗ | NA | NA | NA | NA | NA |
| III.1 | Mucocutaneous bleeding symptoms (epistaxis, hematomas) | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| III.2 | Mucocutaneous bleeding symptoms (epistaxis, hematomas) | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| III.3 | No bleeding symptoms | 4.8, 4.9 | NA | 90, 91 | 360, 275 | 8.98, NA | NA | NA | NA | NA | NA |
| III.4 | Mild mucocutaneous bleeding symptoms | 5.1, 5.8, 5.38 | 16.3, 19.9, 17.4 | 62.6, 76.7, 71.9 | 120, 124, 112 | NR∗ , 12.1, NR∗ | 9.4, 15.7 | 5 μM ADP, 1 μg/mL collagen | 1.7 | Basal: 54 Convulxin: 234 Collagen: 431 | Basal: 86 Convulxin: 96 Collagen: 169 |
| IV.1 | Bleeding after surgery and tooth extraction, oral cavity bleeding, or mucocutaneous bleeding symptoms | 5.5, 5.5 | 15.9, 15.7 | 80.3, 79.4 | 102, 78 | NR∗, 11.9 | 6.9 | 5 μM ADP, 1 μg/mL collagen | 0.7, 0.44, 1.0, 1.2, 1.26 | NA | NA |
| IV.2 | Mild mucocutaneous bleeding symptoms | 5.8, 5.9, 6.2, 6.48 | 15.2, 14.7, 15.6, 17.1 | 68, 68, 79, 75.9 | 135, 125, 84, 84 | NR∗ , NR∗ , 12.8, NR∗ | 13.1, 7.7, 24.1 | 5 μM ADP, 1 μg/mL collagen | 2 | Basal: 51 Convulxin: 125 Collagen: 864 | Basal: 75 Convulxin: 88 Collagen: 155 |
| IV.3 | No bleeding symptoms | 4.6 | 14.0 | 80 | 359 | 8.09 | NA | NA | NA | NA | NA |
| Identity . | Bleeding symptoms . | Erythrocyte count (normal: 4 × 106/μL to 5.2 × 106/μL) . | Red cell distribution width (normal: 11.7% -14.5%) . | Mean corpus volume (normal: 77-95 fL) . | Platelet count (normal: 150 × 109/L to 450 × 109/L) . | Mean platelet volume (normal: 8-12 fL) . | Immature platelet fraction (normal: 1.1% -6.1%) . | Platelet aggregation defect (amplitude <50%) for the indicated agonist . | ATP secretion (normal: 1.2-2.8 μM) . | Flow cytometry for CD62P with MFI basal (normal: 97-196) and after activation with 1 μg/mL convulxin (normal: 305-1606) and 50 μg/mL collagen (normal: 1254-3169) . | Flow cytometry for CD63 with MFI basal (normal: 45-86) and after activation with 1 μg/mL convulxin (normal: 73-121) and 50 μg/mL collagen (normal: 358-507) . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| II.1 | Mild mucocutaneous bleeding symptoms | 4.4 | 17.7 | 88 | 53 | NR∗ | NA | NA | NA | NA | NA |
| II.3 | Mild mucocutaneous bleeding symptoms | 4.5 | 18.5 | 87 | 49 | NR∗ | NA | NA | NA | NA | NA |
| III.1 | Mucocutaneous bleeding symptoms (epistaxis, hematomas) | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| III.2 | Mucocutaneous bleeding symptoms (epistaxis, hematomas) | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| III.3 | No bleeding symptoms | 4.8, 4.9 | NA | 90, 91 | 360, 275 | 8.98, NA | NA | NA | NA | NA | NA |
| III.4 | Mild mucocutaneous bleeding symptoms | 5.1, 5.8, 5.38 | 16.3, 19.9, 17.4 | 62.6, 76.7, 71.9 | 120, 124, 112 | NR∗ , 12.1, NR∗ | 9.4, 15.7 | 5 μM ADP, 1 μg/mL collagen | 1.7 | Basal: 54 Convulxin: 234 Collagen: 431 | Basal: 86 Convulxin: 96 Collagen: 169 |
| IV.1 | Bleeding after surgery and tooth extraction, oral cavity bleeding, or mucocutaneous bleeding symptoms | 5.5, 5.5 | 15.9, 15.7 | 80.3, 79.4 | 102, 78 | NR∗, 11.9 | 6.9 | 5 μM ADP, 1 μg/mL collagen | 0.7, 0.44, 1.0, 1.2, 1.26 | NA | NA |
| IV.2 | Mild mucocutaneous bleeding symptoms | 5.8, 5.9, 6.2, 6.48 | 15.2, 14.7, 15.6, 17.1 | 68, 68, 79, 75.9 | 135, 125, 84, 84 | NR∗ , NR∗ , 12.8, NR∗ | 13.1, 7.7, 24.1 | 5 μM ADP, 1 μg/mL collagen | 2 | Basal: 51 Convulxin: 125 Collagen: 864 | Basal: 75 Convulxin: 88 Collagen: 155 |
| IV.3 | No bleeding symptoms | 4.6 | 14.0 | 80 | 359 | 8.09 | NA | NA | NA | NA | NA |
Boldface indicates values outside the reference range.
ADP, adenosine diphosphate; MFI, mean fluorescent intensity; NA, not applicable; NR, not reported.
NR outside the upper limit of measurement because of the overlap with small erythrocytes.
Blood-derived stem cells from patients IV.2 and III.4 were differentiated into MK, and flow cytometry at day 8 showed no difference in CD41a+/CD42a+ MK compared with the control (supplemental Figure 8). Proplatelets formed in mutant MK were limited to small, straight membrane protrusions that did not evolve further into normally elongated proplatelets, as previously described for stem cell–derived MK from patients with SLFN14 V220D4 (Figure 1E). Interestingly, RNA integrity number (RIN) values determined for RNA from the stem cell–derived MK for patients IV.2 and III.4 also showed lower values compared with RNA from the control on day 8 but not on day 6 of differentiation (Figure 1F), whereas RNA of granulocytes differentiated from the same stem cells showed normal RIN values (supplemental Figure 9).
Defects in megakaryopoiesis using heterozygous and homozygous SLFN14-K219N imMKCL
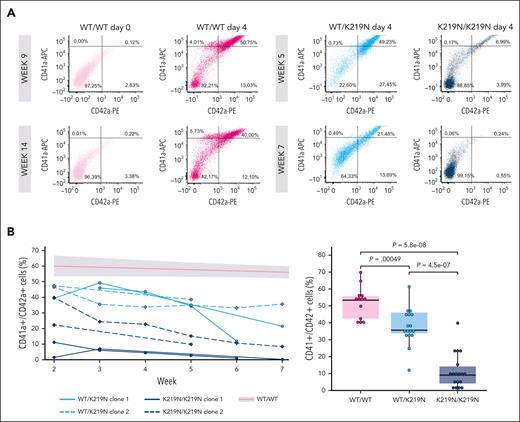
To further explore the effect of SLFN14 K219N on rRNA processing during megakaryopoiesis, we generated 2 heterozygous and 2 homozygous K219N imMKCL using CRISPR/Cas technology and single cell sorting (supplemental Figure 10 top panel). WT (WT/WT), heterozygous (WT/K219N), and homozygous (K219N/K219N) imMKCL cells were differentiated to MK, which were used for functional and morphological studies, as summarized in supplemental Figure 10 bottom panel. Supplemental Figure 11 shows representative images of imMKCL-derived MK during the differentiation of WT/WT cells with the formation of large MK at day 3 and proplatelet formation starting on day 4. CD41a+/CD42a+ MK at day 4 were quantified using flow cytometry after duplicated series of differentiation experiments starting from the 2 clones at different passages (between weeks 2 and 7) (Figure 2A-B). Interestingly, although a severe block in MK differentiation was noted for K219N/K219N cells independent of their passage number, WT/K219N MK showed an increased defect in CD41a+/CD42a+ differentiation by passage number compared with the WT/WT condition (Figure 2B). The passage number showed no effect on CD41a/CD42a expression in nondifferentiated imMKCL cells (supplemental Figure 12), and the relative number of apoptotic MK was comparable for all conditions (supplemental Figure 13). When only day 4 CD41a+/CD42a+ MK at passage week 2 for WT/K219N imMKCL clones was compared with the WT/WT condition, no difference was observed in MK differentiation (supplemental Figure 14), indicating that SLFN14 K219N has a biological effect on imMKCL cells during their culture time. Therefore, week-2 WT/K219N MK are similar to the MK isolated from the patients (supplemental Figure 8).
SLFN14 deficiency results in defective MK differentiation. (A) Example plots of flow cytometry analysis of day 4 differentiated imMKCL cells, indicating the percentage of CD41a+, CD42a+, and CD41a+/CD42a+ MK. WT/WT cells were measured at passage weeks 9 and 14, and WT/K219N and K219N/K219N cells were measured at passage weeks 2 and 7. (B) Percentage of CD41a+/CD42a+ cells over time for 2 clones each of WT/K219N and K219N/K219N cells (left). A trend line with error margins (gray area) was fitted for WT/WT cells measured over 14 weeks in culture and added to the graph. Comparison of mean CD41a+/CD42a+ cells for WT/WT, WT/K219N, and K219N/K219N cells show a significant decrease in double positive cells in SLFN14-deficient cells, as measured during different passage weeks (right). Boxplots represent the middle 50% of the data points, with 25% outliers as whiskers and the mean as a black horizontal line. All individual data points were added as dots to the graph. P values were determined using the pairwise Wilcoxon test.
SLFN14 deficiency results in defective MK differentiation. (A) Example plots of flow cytometry analysis of day 4 differentiated imMKCL cells, indicating the percentage of CD41a+, CD42a+, and CD41a+/CD42a+ MK. WT/WT cells were measured at passage weeks 9 and 14, and WT/K219N and K219N/K219N cells were measured at passage weeks 2 and 7. (B) Percentage of CD41a+/CD42a+ cells over time for 2 clones each of WT/K219N and K219N/K219N cells (left). A trend line with error margins (gray area) was fitted for WT/WT cells measured over 14 weeks in culture and added to the graph. Comparison of mean CD41a+/CD42a+ cells for WT/WT, WT/K219N, and K219N/K219N cells show a significant decrease in double positive cells in SLFN14-deficient cells, as measured during different passage weeks (right). Boxplots represent the middle 50% of the data points, with 25% outliers as whiskers and the mean as a black horizontal line. All individual data points were added as dots to the graph. P values were determined using the pairwise Wilcoxon test.
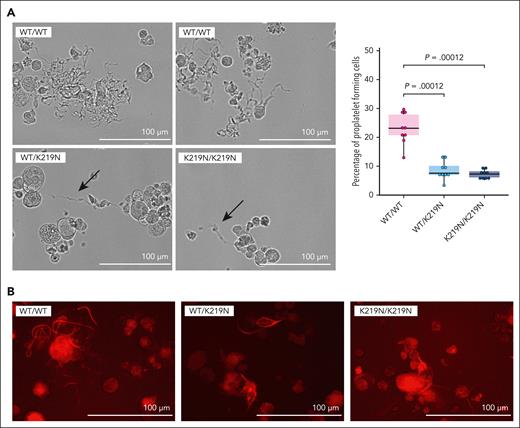
DNA ploidy was studied on day 4 of culture by propidium iodide staining in combination with flow cytometric analysis of CD41a+ MK (supplemental Figure 15). On average, a mean ploidy level of 4.19 ± 0.33 was found for WT/WT MK, 3.47 ± 1.14 for WT/K219N MK, and 2.67 ± 0.32 for K219N/K219N MK. Although a trend toward a lower mean ploidy level for the homozygous condition was observed, this difference was not statistically significant. We also did not observe any differences in DNA ploidy by SiR-DNA staining (supplemental Figure 16). Proplatelet formation was studied by imaging cultured MK on day 5 (triplicate differentiation experiment) (Figure 3A left panel). A significant decrease in the amount of proplatelet-forming MK was observed for WT/K219N and K219N/K219N compared with WT/WT MK (Figure 3A right panel). In addition, proplatelets formed in the mutant MK were limited to small protrusions (Figure 3A-B) as detected in MK cultures from patients (Figure 1E).
SLFN14-deficient MKs display reduced proplatelet formation. (A) Representative brightfield microscopy images illustrating proplatelet formation on differentiation day 5 in WT/WT, WT/K219N, and K219N/K219N cells (left). Proplatelets formed in WT/K219N and K219N/K219N cells are shorter and with a reduced ramification compared with those formed in WT/WT cells (arrows). Imaging was performed on cells at passage week 5 for WT/WT and WT/K219N, and week 3 for K219N/K219N. Quantification of proplatelet-forming cells on day 5 for 3 independent differentiations shows a significant decrease in both WT/K219N and K219N/K219N cells compared with WT/WT (right). On average, 133, 114, and 120 total number of cells were counted for WT/WT, WT/K219N, and K219N/K219N cells, respectively. Statistics performed with a pairwise Wilcoxon test. Boxplots represent the middle 50% of the data points with 25% outliers as whiskers and the mean as a black horizontal line. All individual data points were added as dots to the graph. (B) SiR-tubulin in vivo staining of day 5 differentiated MK shows the presence of shorter proplatelets with reduced ramification for WT/K219N and K219N/K219N conditions compared with WT/WT MK (representative images from a blinded assay). Scale bars, 100 μm.
SLFN14-deficient MKs display reduced proplatelet formation. (A) Representative brightfield microscopy images illustrating proplatelet formation on differentiation day 5 in WT/WT, WT/K219N, and K219N/K219N cells (left). Proplatelets formed in WT/K219N and K219N/K219N cells are shorter and with a reduced ramification compared with those formed in WT/WT cells (arrows). Imaging was performed on cells at passage week 5 for WT/WT and WT/K219N, and week 3 for K219N/K219N. Quantification of proplatelet-forming cells on day 5 for 3 independent differentiations shows a significant decrease in both WT/K219N and K219N/K219N cells compared with WT/WT (right). On average, 133, 114, and 120 total number of cells were counted for WT/WT, WT/K219N, and K219N/K219N cells, respectively. Statistics performed with a pairwise Wilcoxon test. Boxplots represent the middle 50% of the data points with 25% outliers as whiskers and the mean as a black horizontal line. All individual data points were added as dots to the graph. (B) SiR-tubulin in vivo staining of day 5 differentiated MK shows the presence of shorter proplatelets with reduced ramification for WT/K219N and K219N/K219N conditions compared with WT/WT MK (representative images from a blinded assay). Scale bars, 100 μm.
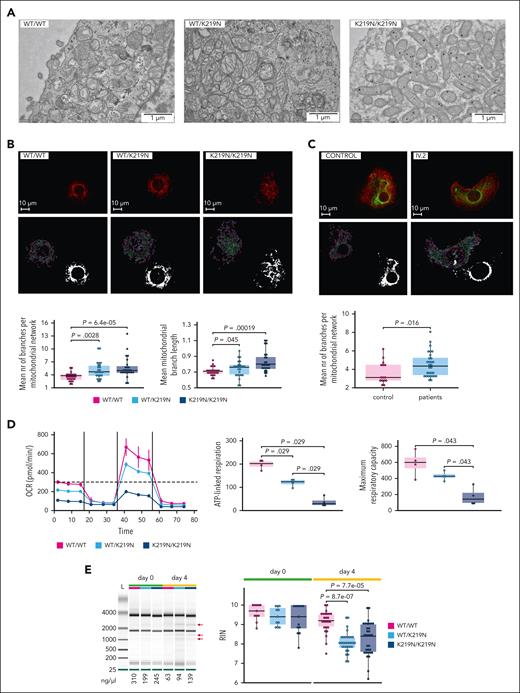
EM imaging of day 3 MK was performed blinded to study the intracellular organization. Both WT/K219N and K219N/K219N displayed lysosomal inclusions and multilamellar vesicles, which were absent in the WT/WT MK (supplemental Figure 17). In addition, abnormally large or elongated mitochondria were observed (Figure 4A; supplemental Figure 17). Therefore, we further analyzed mitochondrial structures in day 3 MK using confocal imaging of MitoTracker Red staining (Figure 4B). Mitochondrial Network Analysis software was used to quantify differences in mitochondrial morphology.15 Though the total mitochondrial volume (footprint) was similar in MK for all conditions (supplemental Figure 18), a significant increase in the number of branches per mitochondrial network and the mean branch length was observed for WT/K219N and K219N/K219N MK (Figure 4B bottom panels). The Mitochondrial Network Analysis of ATP5E-stained mitochondria in MK from patients IV.2 and III.4 also showed a significant increase in the number of branches per mitochondrial network (Figure 4C), while the other parameters were normal (data not shown). The mitochondrial OCR was measured in MK on day 4 for 4 differentiation experiments using the Seahorse assay (Figure 4D left panel). The maximum respiratory capacity and ATP-linked respiration were significantly downregulated in WT/K219N and K219N/K219N MK (Figure 4D right panels; supplemental Figure 19 upper panel), whereas the other parameters showed a trend toward impairment, but these values were not significant (supplemental Figure 19 bottom panels).
SLFN14-deficient MKs show a mitochondrial defect. (A) Transmission EM imaging of differentiation day 3 MK cells shows abnormal mitochondria in SLFN14-deficient MK compared with WT/WT MK. Images taken after a blinded analysis of MK at passage week 17 for WT/WT and WT/K219N and week 3 for K219N/K219N. (B) Confocal imaging of day 4 MK cells from WT and mutant imMKCL with MitoTracker Red to visualize (top) and quantify (bottom) mitochondria. Imaging performed at passage week 6 for WT/K219N and K219N/K219N, and week 13 for WT/WT. Quantification of the mean number (nr) of branches per mitochondrial network (bottom left) and mean mitochondrial branch length (bottom right) as calculated by the Mitochondrial Network Analysis software for 25 randomly selected MK containing a single nucleus for each condition. P values were determined using the pairwise Wilcoxon test for all comparisons. (C) Confocal imaging of day 8 MK from blood-derived stem cells using the ATP5E antibody (green) to visualize (top) and quantify (bottom) mitochondria. Phalloidin staining in red. Quantification of the mean number of branches per mitochondrial network (bottom) as calculated by the Mitochondrial Network Analysis software for 25 randomly selected MK containing a single nucleus for each condition. P values were determined using the Wilcoxon test. (D) OCR measurements were obtained over time (minutes) using the extracellular flux analyzer from Seahorse Bioscience for WT/WT, WT/K219N, and K219N/K219N MK on day 4 (left). Values represent the mean and standard deviation for each condition of the Seahorse assays obtained for 4 replicates from 2 differentiation experiments. The mitochondrial stress test was used to obtain diverse parameters: ATP-linked OCR (by adding the ATP synthase inhibitor oligomycin), maximal OCR (by adding the uncoupling agent FCCP), and complete inhibition of the mitochondria (by adding the inhibitor antimycin) (right top). The maximal respiration capacity and ATP-linked respiration were significantly reduced for WT/K219N and K219N/K219N MK compared with WT/WT MK (right). Statistics using a pairwise Wilcoxon test was performed. Boxplots in all panels represent the middle 50% of the data points with, 25% outliers as whiskers and the mean as a black horizontal line. All individual data points were added as dots to the graph. (E) Representative gel electrophoresis image showing rRNA degradation fragments (red arrows) for day 4 differentiated WT/K219N and K219N/K219N MK that are not present in WT/WT MK or undifferentiated cells (left). Experiments performed at passage week 5 for day 0 and week 7 for day 4. The concentration of total RNA (ng/μL) for the different samples was added below the figure. Quantification of the RIN values obtained for RNA samples of imMKCL cells from 4 (day 0) and 9 (day 4) independent differentiation experiments on days 0 and 4 WT/WT, WT/K219N, and K219N/K219N MK (multiple technical replicates included per time point per condition) (right). Experiments performed at passage weeks 2, 3, 4, 5, 7, 9, 12, 23, and 25. Boxplots represent the middle 50% of the data points with, 25% outliers as whiskers and the mean as a black horizontal line. All individual data points were added as dots to the graph. Statistical analyses were performed using the pairwise Wilcoxon test for all comparisons. Scale bars, 1 μm (A) and 10 μm (B-C).
SLFN14-deficient MKs show a mitochondrial defect. (A) Transmission EM imaging of differentiation day 3 MK cells shows abnormal mitochondria in SLFN14-deficient MK compared with WT/WT MK. Images taken after a blinded analysis of MK at passage week 17 for WT/WT and WT/K219N and week 3 for K219N/K219N. (B) Confocal imaging of day 4 MK cells from WT and mutant imMKCL with MitoTracker Red to visualize (top) and quantify (bottom) mitochondria. Imaging performed at passage week 6 for WT/K219N and K219N/K219N, and week 13 for WT/WT. Quantification of the mean number (nr) of branches per mitochondrial network (bottom left) and mean mitochondrial branch length (bottom right) as calculated by the Mitochondrial Network Analysis software for 25 randomly selected MK containing a single nucleus for each condition. P values were determined using the pairwise Wilcoxon test for all comparisons. (C) Confocal imaging of day 8 MK from blood-derived stem cells using the ATP5E antibody (green) to visualize (top) and quantify (bottom) mitochondria. Phalloidin staining in red. Quantification of the mean number of branches per mitochondrial network (bottom) as calculated by the Mitochondrial Network Analysis software for 25 randomly selected MK containing a single nucleus for each condition. P values were determined using the Wilcoxon test. (D) OCR measurements were obtained over time (minutes) using the extracellular flux analyzer from Seahorse Bioscience for WT/WT, WT/K219N, and K219N/K219N MK on day 4 (left). Values represent the mean and standard deviation for each condition of the Seahorse assays obtained for 4 replicates from 2 differentiation experiments. The mitochondrial stress test was used to obtain diverse parameters: ATP-linked OCR (by adding the ATP synthase inhibitor oligomycin), maximal OCR (by adding the uncoupling agent FCCP), and complete inhibition of the mitochondria (by adding the inhibitor antimycin) (right top). The maximal respiration capacity and ATP-linked respiration were significantly reduced for WT/K219N and K219N/K219N MK compared with WT/WT MK (right). Statistics using a pairwise Wilcoxon test was performed. Boxplots in all panels represent the middle 50% of the data points with, 25% outliers as whiskers and the mean as a black horizontal line. All individual data points were added as dots to the graph. (E) Representative gel electrophoresis image showing rRNA degradation fragments (red arrows) for day 4 differentiated WT/K219N and K219N/K219N MK that are not present in WT/WT MK or undifferentiated cells (left). Experiments performed at passage week 5 for day 0 and week 7 for day 4. The concentration of total RNA (ng/μL) for the different samples was added below the figure. Quantification of the RIN values obtained for RNA samples of imMKCL cells from 4 (day 0) and 9 (day 4) independent differentiation experiments on days 0 and 4 WT/WT, WT/K219N, and K219N/K219N MK (multiple technical replicates included per time point per condition) (right). Experiments performed at passage weeks 2, 3, 4, 5, 7, 9, 12, 23, and 25. Boxplots represent the middle 50% of the data points with, 25% outliers as whiskers and the mean as a black horizontal line. All individual data points were added as dots to the graph. Statistical analyses were performed using the pairwise Wilcoxon test for all comparisons. Scale bars, 1 μm (A) and 10 μm (B-C).
RNA samples from day 4 MK for WT/K219N and K219N/K219N, analyzed with the Agilent 2100 BioAnalyzer, showed rRNA degradation fragments that were not present in WT/WT MK (Figure 4E left panel). In agreement with this, the RIN values for these samples were significantly decreased at day 4 but not at day 0 (Figure 4E right panel). RNA samples from day 6 MK showed that rRNA degradation bands increased in intensity during differentiation, and these were associated with lower RIN values and 28S rRNA peaks (supplemental Figure 20). These experiments indicate that rRNA degradation is initiated only during megakaryopoiesis and in the presence of SLFN14 K219N, suggesting gain-of-function activity for this mutant. This hypothesis is supported by the normal phenotype present in SLFN14 depleted MK. Day 4 MK from imMKCL cells with a nearly 50% reduction in SLFN14 levels showed normal RIN values compared with cells treated with control RNA interference (supplemental Figure 21).
RNA-seq shows enhanced translation and transcription pathways in platelets from patients with K219N
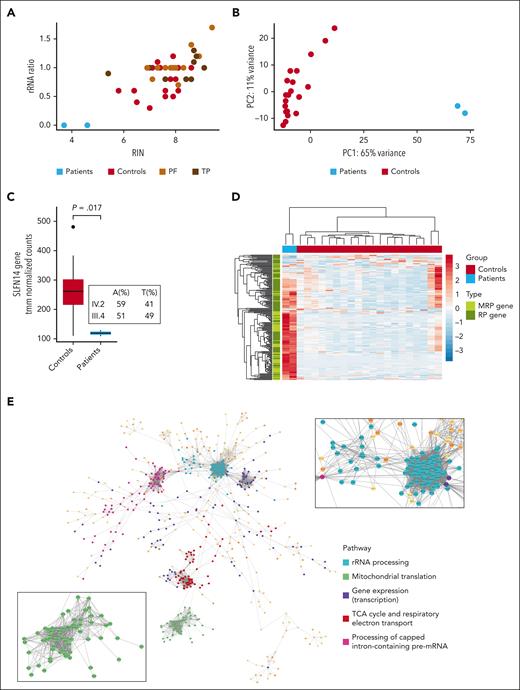
Platelets were isolated from the blood of patients IV.2 and III.4 and 19 unrelated healthy donors for RNA extraction. RIN values and rRNA ratios, determined using the 2200 TapeStation system, showed a distinct reduction in the RNA samples from both patients compared with the values for all controls (Figure 5A). Interestingly, their RIN and rRNA ratios were also lower than those detected in RNA samples from platelets of unrelated patients diagnosed with either a platelet function defect or TP, but without a known genetic cause (Figure 5A). Sample traces shown in supplemental Figure 22 for RNA samples of the patients vs those from 2 randomly selected controls illustrate the presence of reduced 28S and 18S rRNA peaks in combination with rRNA degradation fragments in samples from the patients. These RNA samples were used for total RNA-seq and differential gene expression analysis. Principal component analysis performed for the top 500 genes with the highest variance across all samples showed a clear distinction between patients and controls according to the first principal component, which accounted for 65% of all variance in the data set, illustrating the extensive impact of the SLFN14 K219N variant on the platelet transcriptome (Figure 5B). In line with reduced SLFN14 protein levels, decreased SLFN14 mRNA (with a similar frequency of the mutant [T] vs WT [A] allele) was detected in platelets from the patients compared with those from healthy controls (Figure 5C). Differential gene expression analysis yielded 2999 and 2888 significantly (|log2 fold change| >1, false discovery rate <0.05) up- and downregulated genes, respectively, in patients compared with controls (supplemental Table 1A; supplemental Figure 23). Remarkably, these downregulated genes were not enriched in any biological pathway. Interestingly, the genes for integrin ɑIIbβ3 (ITGA2B and ITGB3) were significantly downregulated in platelets from these patients, in parallel with their low CD41a expression (supplemental Figures 23 and 2). The upregulated genes were significantly enriched in pathways involved in (mitochondrial) translation and transcription (supplemental Table 1B), with a significant upregulation of 134 ribosomal protein genes (RPGs). Figure 5D shows a heatmap representing tmm-normalized counts for all expressed (>10 reads on average in controls) RPGs and mitochondrial RPGs. In total, we found that 50 and 38 large subunits of RPGs and mitochondrial RPGs, respectively, and 27 and 19 small subunits of RPGs and mitochondrial RPGs, respectively, were significantly upregulated in platelets from patients, indicating a common pathway that stimulates (mitochondrial) RPG expression, such as the mammalian target of rapamycin complex 1 (mTORC1) pathway.16,17 To better visualize the molecular pathways connecting the function of the detected upregulated genes, a network of STRING protein-protein interactions was constructed using Cytoscape3. Only connections with a confidence score >99% were considered when building the network, and only the largest connected network was used for the analysis. Pathway analysis of the network showed enrichment for rRNA processing, mitochondrial translation, gene expression, respiratory electron transport, and pre-mRNA processing (Figure 5E).
Platelet RNA-seq analysis shows extensive gene expression differences with upregulation of translation and transcription pathways in SLFN14 K219N carriers. (A) Dotplot showing the RIN value vs rRNA ratio of platelet RNA-seq samples from SLFN14 patients IV.2 and III.4, 19 healthy controls, and 26 unrelated, undiagnosed patients with a platelet function (PF) defect or TP. (B) Principal component (PC) analysis plot based on the top 500 genes with the highest variance among samples. (C) Boxplot showing tmm-normalized counts for SLFN14 in patients IV.2 and III.4 and 20 controls as detected by total RNA-seq. Boxplots represent the middle 50% of the data points with, 25% outliers as whiskers and the mean as a black horizontal line. (D) Heatmap showing tmm-normalized counts for all expressed (>10 reads on average in the control group) mitochondrial (M) RPGs and RPGs in patients IV.2 and III.4 and controls. Statistics were performed using the Wilcoxon test. (E) A gene-gene association network for all significantly upregulated genes (log2 fold change >1, false discovery rate <0.05) in platelets from patients IV.2 and III.4, created in Cytoscape. Five subnetworks among the 2762 nodes were identified in the largest connected network. The zoom-in images show the mitochondrial RPGs (green) and RPGs (light blue). TCA, tricarboxylic acid.
Platelet RNA-seq analysis shows extensive gene expression differences with upregulation of translation and transcription pathways in SLFN14 K219N carriers. (A) Dotplot showing the RIN value vs rRNA ratio of platelet RNA-seq samples from SLFN14 patients IV.2 and III.4, 19 healthy controls, and 26 unrelated, undiagnosed patients with a platelet function (PF) defect or TP. (B) Principal component (PC) analysis plot based on the top 500 genes with the highest variance among samples. (C) Boxplot showing tmm-normalized counts for SLFN14 in patients IV.2 and III.4 and 20 controls as detected by total RNA-seq. Boxplots represent the middle 50% of the data points with, 25% outliers as whiskers and the mean as a black horizontal line. (D) Heatmap showing tmm-normalized counts for all expressed (>10 reads on average in the control group) mitochondrial (M) RPGs and RPGs in patients IV.2 and III.4 and controls. Statistics were performed using the Wilcoxon test. (E) A gene-gene association network for all significantly upregulated genes (log2 fold change >1, false discovery rate <0.05) in platelets from patients IV.2 and III.4, created in Cytoscape. Five subnetworks among the 2762 nodes were identified in the largest connected network. The zoom-in images show the mitochondrial RPGs (green) and RPGs (light blue). TCA, tricarboxylic acid.
Heterozygous and homozygous SLFN14-K219N MKs show enhanced ribosome biogenesis via the mTORC1 pathway
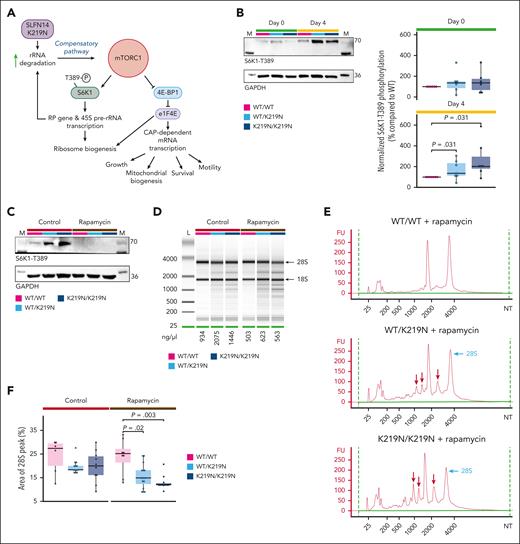
We hypothesized that rRNA degradation is followed by a compensatory mechanism that initiates ribosomal biogenesis via mTORC1 signaling, as described for human cells with defective rRNA processing.18 The mTORC1 positively regulates the transcription of multiple mitochondrial RPGs.19 Such a mechanism could explain the elevated mitochondrial RPGs detected in platelet RNA-seq data but also the minimal effect on the rRNA ratio as rRNA degradation is accompanied by increased rRNA production. The pathway proposed in Figure 6A illustrates how elevated mTORC1 signaling can explain many of the upregulated pathways detected in the platelet RNA-seq data, such as rRNA and pre-mRNA processing, gene expression, mitochondrial translation, and respiratory electron transport. Activated mTORC1 is known to phosphorylate and activate ribosomal protein S6 kinase 1 (S6K1) to initiate ribosome biogenesis (Figure 6A).20 Therefore, we studied S6K1-T389 phosphorylation during megakaryopoiesis. Interestingly, WT/K219N and K219N/K219N MK showed higher S6K1-T389 phosphorylation levels than WT/WT MK (Figure 6B), but only on day 4, whereas these differences were not observed in undifferentiated cells. Although protein expression of SLFN14 in patient platelets was reduced (Figure 1D), no significant difference was detected in undifferentiated cells or day 4 MK at the protein level (supplemental Figure 24).
Overactivation of mTORC1 pathway acts as a compensatory mechanism for rRNA degradation. (A) Schematic overview of the mTORC1 pathway. We hypothesized that increased rRNA degradation mediated by SLFN14 K219N stimulates mTORC1 signaling as a compensatory mechanism. Active mTORC1 signaling stimulates ribosomal biogenesis via phosphorylation of S6K1-T389. In addition, mTORC1 is involved in cell growth, survival, motility, and mitochondrial biogenesis. Figure created using BioRender.com. (B) Representative immunoblots showing phosphorylated S6K1-T389 protein expression in undifferentiated cells and day 4 MK for WT/WT, WT/K219N, and K219N/K219N conditions (left). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the loading control for normalization. Quantification of immunoblot data for 6 (day 0) and 7 (day 4) independent differentiation experiments show a significant increase in S6K1-T389 phosphorylation in WT/K219N and K219N/K219N day 4 MK compared with WT/WT, whereas no difference was seen in undifferentiated cells (day 0). Analysis performed on cells between passage weeks 10 to 13 for WT/WT and between weeks 3 and 6 for WT/K219N and K219N/K219N (right). Statistics were performed with a pairwise Wilcoxon test for all comparisons. (C) Immunoblot analysis showing a complete lack of S6K1-T389 phosphorylated protein in day 4 MK treated with rapamycin (and without) at passage week 5. GAPDH was used as a loading control. (D) Gel electrophoresis analysis shows rapamycin treatment exacerbates rRNA degradation in SLFN14 K219N cells. Analysis of cells at passage week 5. (E) Electrophoresis traces of rapamycin-treated cells show increased RNA degradation fragment peaks (red arrows) and reduced 28S peaks (light blue arrows) in WT/K219N cells and an even stronger effect in K219N/K219N day 4 MK. Analysis performed on cells at passage week 5. (F) Treatment with rapamycin significantly reduces the area of the 28S peak for both WT/K219N and K219N/K219N on day 4 MK. Analysis performed on cells between passage weeks 3 to 5. Statistics were performed with a pairwise Wilcoxon test for all comparisons. Boxplots for all panels represent the middle 50% of the data points, with 25% outliers as whiskers and the mean as a black horizontal line. All individual data points were added as dots to the graph. FU, fluorescence units; NT, nucleotides.
Overactivation of mTORC1 pathway acts as a compensatory mechanism for rRNA degradation. (A) Schematic overview of the mTORC1 pathway. We hypothesized that increased rRNA degradation mediated by SLFN14 K219N stimulates mTORC1 signaling as a compensatory mechanism. Active mTORC1 signaling stimulates ribosomal biogenesis via phosphorylation of S6K1-T389. In addition, mTORC1 is involved in cell growth, survival, motility, and mitochondrial biogenesis. Figure created using BioRender.com. (B) Representative immunoblots showing phosphorylated S6K1-T389 protein expression in undifferentiated cells and day 4 MK for WT/WT, WT/K219N, and K219N/K219N conditions (left). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the loading control for normalization. Quantification of immunoblot data for 6 (day 0) and 7 (day 4) independent differentiation experiments show a significant increase in S6K1-T389 phosphorylation in WT/K219N and K219N/K219N day 4 MK compared with WT/WT, whereas no difference was seen in undifferentiated cells (day 0). Analysis performed on cells between passage weeks 10 to 13 for WT/WT and between weeks 3 and 6 for WT/K219N and K219N/K219N (right). Statistics were performed with a pairwise Wilcoxon test for all comparisons. (C) Immunoblot analysis showing a complete lack of S6K1-T389 phosphorylated protein in day 4 MK treated with rapamycin (and without) at passage week 5. GAPDH was used as a loading control. (D) Gel electrophoresis analysis shows rapamycin treatment exacerbates rRNA degradation in SLFN14 K219N cells. Analysis of cells at passage week 5. (E) Electrophoresis traces of rapamycin-treated cells show increased RNA degradation fragment peaks (red arrows) and reduced 28S peaks (light blue arrows) in WT/K219N cells and an even stronger effect in K219N/K219N day 4 MK. Analysis performed on cells at passage week 5. (F) Treatment with rapamycin significantly reduces the area of the 28S peak for both WT/K219N and K219N/K219N on day 4 MK. Analysis performed on cells between passage weeks 3 to 5. Statistics were performed with a pairwise Wilcoxon test for all comparisons. Boxplots for all panels represent the middle 50% of the data points, with 25% outliers as whiskers and the mean as a black horizontal line. All individual data points were added as dots to the graph. FU, fluorescence units; NT, nucleotides.
To further investigate the compensatory mTORC1 pathway in SLFN14-deficient MK, we treated the cells with the mTORC1 inhibitor rapamycin after 24 hours of differentiation. The addition of rapamycin completely blocks S6K1-T389 phosphorylation under all conditions (Figure 6C; supplemental Figure 25). Flow cytometry analysis of rapamycin-treated day 4 mutant MK showed a significant decrease in MK differentiation compared that to in WT/WT MK (supplemental Figure 26). Although a reduction in CD41a+/CD42a+ MK in rapamycin-treated compared with nontreated WT/K219N was present, this difference was not significant after multiple testing corrections. We hypothesized that mTORC1 inhibition would block the ribosomal biogenesis pathway, exacerbating the RNA degradation profile observed in WT/K219N and K219N/K219N MK compared with that in WT/WT MK. Indeed, day 4 MK treated with rapamycin showed more intense degradation fragments for WT/K219N and K219N/K219N conditions compared with WT/WT (Figure 6D-E). Interestingly, a significant decrease in the area of the 28S peak (Figure 6F) was observed for WT/K219N and K219N/K219N day 4 MK compared with that in WT/WT MK, as determined in 3 independent differentiation experiments.
Discussion
The heterozygous K219N SLFN14 variant causes mild TP with minor platelet function defects, whereas patients have an obvious bleeding tendency.2,4 We performed platelet RNA-seq in SLFN14-deficient patients. Unexpectedly, a high number of significantly up- and downregulated genes (2999 and 2888, respectively) were detected. For comparison, RNA-seq of platelets from patients with TP with a pathogenic variant in the transcription factor IKZF5 resulted in a total of 1194 differentially expressed genes.13 In contrast, RNA-seq of platelets from patients with TP with a pathogenic variant in NBEAL2 detected a total of only 95 differentially expressed genes.21 Notably, these 2 studies used the same platelet isolation protocol used in this study. This might indicate that TP is not a major determining factor for an altered gene expression pattern, whereas defects in transcription regulation, as expected, result in a high number of differentially expressed genes. This suggests that SLFN14 defects have a major effect on gene expression regulation in platelets, beyond the regulation of transcription factor. The upregulated genes were enriched in rRNA processing, mitochondrial translation, gene expression, respiratory electron transport, and pre-mRNA processing. Such pathways predict major effects on gene expression.
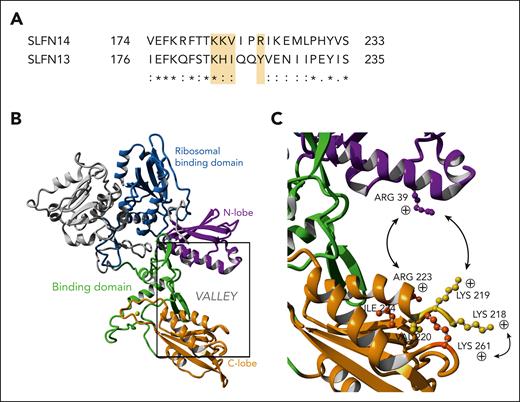
The 4 missense variants that cause TP in humans are located in the N-terminal AAA domain of SLFN14, which is present in all SLFN proteins, and was shown to be homologous to other ATP/guanosine triphosphate binding domains that function in DNA and RNA metabolism.3 A recent 3-dimensional structure generated for rat SLFN13 shows how a valley is created between the N-terminal and C-terminal lobes, and its width of ∼23 Å is exactly suitable for clamping base-paired RNAs.22 They showed that SLFN13 functions as an RNase able to digest transfer RNAs and rRNAs and that it can inhibit protein synthesis in cells; however, in contrast to some other RNases, it functions as a monomer. The valley contains positively charged amino acids (in C-terminal lobe: Lys38, Arg39, and Lys42 vs in N-terminal lobe: Arg217, Lys224, and Lys276) that upon substitution to alanine resulted in reduced nucleolytic activity, indicating that changing the valley width can change the RNase activity.22 Compared with rat SLFN13, human SLFN14 shows a very high sequence consistency in the positively charged C- and N-terminal lobes (Figure 7A). In addition, rabbit SLFN14 has been reported to be active in transfer RNA/rRNA cleavage.7 Pathogenic SLFN14 missense variants are all located in the N-terminal lobe and are predicted to alter the positively charged region or its confirmation within this lobe (Figure 7B). This would result in a change in the valley width and subsequently alter the RNase activity. RNase activity measurements for rat SLFN13 were performed using a recombinant shorter protein fragment (lacking the SWADL and helicase domains) and only substitutions to alanine were used.7 Though most SLFN13 mutants showed loss of RNase activity, some of the mutants described in the supplemental data of the study showed a gain-of-function activity, but only in vitro assays were used. Here, we detected enhanced rRNA cleavage in differentiated heterozygous and homozygous K219N MK using genetically modified imMKCL and in MK, and platelets from patients carrying this variant. Interestingly, nondifferentiated heterozygous and homozygous K219N imMKCL cells did not show this rRNA degradation pattern. The complexity of this lineage-specific phenotype makes it very difficult to study the mutant SLFN14 RNase activity using in vitro assays or as previously shown, using transfected cells.5 Indeed, overexpression of mutant SLFN14 in DAMI and HEK293T cells had a similar effect on ribosomal binding and rRNA degradation as WT SLFN14.5 Probably, our more physiological cell model for MK and patient-derived platelets was able to detect this difference in RNase activity. The study from Mills et al showed that ribosome homeostasis is differently regulated in enucleated platelets and reticulocytes when compared with nucleated cells.23 They reason that this difference is relevant to understand how genetic defects that cause ribosomopathies often only affect hematopoietic cells. Such a lineage-specific defect could explain why patients with SLFN14 variants primarily present TP. Because we also detected a minor defect in the erythrocytes of our patients, it would be interesting to further study the RNase activity of mutant SLFN14 during erythropoiesis. Although rRNA degradation fragments were detected in SLFN14-defective MK and platelets, 28S and 18S rRNA peaks were not reduced. This can be explained by the fact that rRNA degradation is followed by a compensatory mechanism that initiates ribosomal biogenesis via mTORC1 signaling, as described for other defects in rRNA processing.18 Indeed, treatment of imMKCL cells with the mTORC1 inhibitor rapamycin during differentiation enhances rRNA degradation with reduced 28S rRNA peaks in SLFN14-defective MK. This is because of the loss of a compensatory pathway that stimulates ribosomal biogenesis. mTORC1 signaling has been described as a major regulator of mitochondrial and lysosomal functions, the 2 major phenotypic cellular alterations present in SLFN14-defective MK.24,25 We found enlarged mitochondrial networks and elongated mitochondria in SLFN14-deficient MK that were associated with reduced OCR. Interestingly, MK-specific knockout mice for the mitochondrial fission regulator Dynamin-like protein 1 present with mild macrothrombocytopenia and hyperextended mitochondrial networks.26 Dynamin-like protein 1 inhibition in cancer cells has been associated with both reduced and increased OCR, indicating that the link between mitochondrial fission and fusion mechanisms and mitochondrial metabolism is very complex.27 It is known that mTORC1 can interfere with these processes.28
Variants in positively charged patches flanking the binding valley of SLFN14 result in altered protein function. (A) Sequence homology between human SLFN14 and SLFN13 shows conservation of positively charged amino acid side chains for known TP-related variants (yellow highlight). “∗” indicates a fully conserved residue; “:” a conservation between groups of strongly similar properties, scoring >0.5 in the Gonnet PAM 250 matrix; and “.” a conservation between groups of weakly similar properties, scoring ≤0.5 in the Gonnet PAM 250 matrix. (B) Overview of the complete SLFN14 monomer (P0C7P3). Model based on known SLFN13 protein structure. Important function regions have been highlighted. (C) Closeup view of the valley region of SLFN14, containing positively charged residues with known variants related to TP (K218, K219, V220, and R223).
Variants in positively charged patches flanking the binding valley of SLFN14 result in altered protein function. (A) Sequence homology between human SLFN14 and SLFN13 shows conservation of positively charged amino acid side chains for known TP-related variants (yellow highlight). “∗” indicates a fully conserved residue; “:” a conservation between groups of strongly similar properties, scoring >0.5 in the Gonnet PAM 250 matrix; and “.” a conservation between groups of weakly similar properties, scoring ≤0.5 in the Gonnet PAM 250 matrix. (B) Overview of the complete SLFN14 monomer (P0C7P3). Model based on known SLFN13 protein structure. Important function regions have been highlighted. (C) Closeup view of the valley region of SLFN14, containing positively charged residues with known variants related to TP (K218, K219, V220, and R223).
In conclusion, we identified ribosomal biogenesis via mTORC1 signaling as an important regulator of megakaryopoiesis based on observations in patients with SLFN14-related TP.
Acknowledgments
This work was supported by grants from the KU Leuven BOF (Belgium) (grant C14/19/096) and FWO (Belgium) (grants G072921N and 1115222N) and research grants from Novo Nordisk (Denmark), Bayer (Germany), CSL Behring (King of Prussia, PA), and Swedish Orphan Biovitrum AB (Sweden).
Authorship
Contribution: F.V.D. performed experiments, data analysis, and wrote the manuscript; K.R. and C.T. performed immortalized megakaryocyte cell line experiments; C.V.L., V.L., C.V.G., and K.P. studied the patients and collected the clinical data; K.E. provided the differentiation protocol and immortalized megakaryocyte cell line model; K.F. designed the study and cowrote the manuscript; and all authors edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Kathleen Freson, Department of Cardiovascular Sciences, Center for Molecular and Vascular Biology, University of Leuven, UZ Herestr 49–bus 911, 3000 Leuven, Belgium; e-mail: kathleen.freson@kuleuven.be.
References
Author notes
Platelet RNA sequencing data reported in this article have been deposited in the European Genome-phenome Archive at https://ega-archive.org (accession number EGAS00001006339).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal