In this issue of Blood, Ver Donck et al,1 investigated a dominantly inherited platelet defect with a mild-to-moderate bleeding diathesis in a family due to a mutation in SLFN14,1,2 which is involved in RNA degradation, especially ribosomal RNA (rRNA) but also transfer RNA (tRNA) and messenger RNA (mRNA).2,3 The authors demonstrate a loss of rRNA in platelets in affected family members and studied the other consequences of this mutation.1 These findings offer insights into how a potential endoribonuclease SLFN14 may affect megakaryocyte development, and have pleiotropic effects on both megakaryocytes and platelets. The study also suggests a potential therapeutic target for these patients, with implications for understanding the differentiation of large, polyploidy megakaryocytes. Overall, the molecular bases of inherited platelet disorders continue to provide valuable insights into megakaryocyte and platelet biology.
The authors’ studies used patient platelets and megakaryocytes and CRISPR/Cas9–engineered cells from an immortalized megakaryocyte progenitor cell line (imMKCL) to define the consequences of the K219N amino acid substitution in SLFN14.1 imMKCL is an immortalized induced pluripotent stem cell (iPSC) intermediate line that, upon withdrawal of doxycycline, completes its differentiation into megakaryocyte-like cells.4,5 Overall, studies using the primary cells and the imMKCL were consistent in terms of morphologic changes in the megakaryocytes.4,5 Further studies of the imMKCL demonstrated multiple, apparently downstream, pleiotropic effects of the SLFN14K219N mutation on differentially expressed genes in the mutated imMKCL.1 Of particular importance was the enhancement of genes involved in rRNA processing, which may explain why the levels of 28S and 18S rRNAs in the mutant imMKCL megakaryocytes were normal yet associated with enhanced levels of degraded rRNA bands. Furthermore, mitochondrial translation and protein expression pathways were enhanced in the mutant imMKCL, consistent with the mitochondrial morphologic abnormalities observed via electron microscopy and the marked reduction in mitochondrial oxygen consumption rate by mutated imMKCL-derived megakaryocytes.1
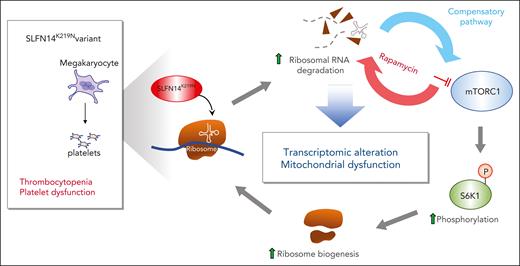
The authors point out that the changes in both ribosomal and mitochondrial biogenesis suggest that the SLFN14K219N mutation enhances the mammalian target of rapamycin complex 1 (mTORC1) pathway. To test this, the authors treated the differentiating imMKCL megakaryocytes with rapamycin to inhibit mTORC1 pathway activity and saw decreased megakaryocyte differentiation concomitant with decreased levels of 28S rRNA and markedly enhanced levels of rRNA degradation fragments, supporting the model proposed in the figure.
Proposed pathways underlying the phenotype of the SLFN14K219N variant. SLFN14K219N mutant leads to degradation of rRNA accompanied with compensatory activation of the mTORC1 pathway via phosphorylation of the ribosomal protein S6 kinase 1 (S6K1) at threonine 389 and enhanced ribosomal biogenesis. Inhibition of the mTORC1 pathway by rapamycin blocks ribosomal protein and increases degradation of rRNA. mTORC1 activation also leads to transcriptomic alteration and mitochondrial stress, resulting in thrombocytopenia and platelet dysfunction.
Proposed pathways underlying the phenotype of the SLFN14K219N variant. SLFN14K219N mutant leads to degradation of rRNA accompanied with compensatory activation of the mTORC1 pathway via phosphorylation of the ribosomal protein S6 kinase 1 (S6K1) at threonine 389 and enhanced ribosomal biogenesis. Inhibition of the mTORC1 pathway by rapamycin blocks ribosomal protein and increases degradation of rRNA. mTORC1 activation also leads to transcriptomic alteration and mitochondrial stress, resulting in thrombocytopenia and platelet dysfunction.
Thus, the authors have shown that the K219N mutation in the endoribonuclease SLFN14 appears to involve rRNA degradation and alterations in the mTORC1 pathway (see figure), although other pathways may be affected in the developing megakaryocytes because there were robust changes in many important intracellular pathways. Why the degradation of rRNAs leads to such widespread effects, whether the endoribonuclease activity of SLFN14 targeting rRNAs accounts for all the pathology because SLFN14 may also target tRNAs and mRNAs, and whether these, as yet unidentified tRNAs or mRNAs, may contribute to the observed phenotype remain open questions.
Another area of future effort would be to understand the basis of the rRNA degradation. SLFN14 K218N results in a lower overall SLFN14 mRNA level with both the wildtype (WT) and the mutant mRNAs being reduced. The mutation also results in a decreased overall level of SLFN14. How this reduction in SLFN14 is linked to an increase in the ribonuclease activity, suggested by the present study, needs further explanation.
Many of the studies by the authors used imMKCL megakaryocytes that had been gene edited to replicate the heterozygote SLFN14WT/K219N phenotype observed in patients and create cells that were homozygous for the mutant SLFN14.1 There are limitations of these cells. One variance of the imMKCL studies from studies of patient primary platelets was the marked decrease in rRNA in the patient platelets but normal or near-normal rRNA levels even in the homozygous mutant imMKCL-derived megakaryocytes. Perhaps the half-life of rRNA is shortened and leads to low levels in the patients’ anucleate circulating platelets, whereas imMKCL-derived megakaryocytes replenish the rRNA pool by compensatory activation of the mTORC1 pathway so that it required treating these megakaryocytes with rapamycin to observe a decrease in 28S rRNA1 (see figure).
In addition, imMKCL cells are already intermediately differentiated to megakaryocytes.4,5 It was clear that the heterozygous imMKCL-SLFN14WT/K219N lines were significantly affected by cell passage, with markedly reduced terminal megakaryocyte production even with 4 passages. Whether studies beginning with iPSCs rather than intermediate imMKCs would have more fully replicated primary hematopoietic progenitor cells and give a fuller picture of events during megakaryopoiesis is unclear but should be considered in future gene mutation studies of SLFN14 or other genes that may have an affect early in megakaryocyte differentiation.
Finally, why megakaryocytes are uniquely affected in this disorder is unaddressed. Clearly, a number of systemic disorders affecting RNA-processing steps, such as RNA splicing and ribosomal function,6-8 target hematopoietic lineages, especially the related lineages of megakaryocytes and erythrocytes. These 2 lineages are closely linked in differentiation,9,10 and, perhaps, this linkage or these lineages’ dependency on extreme gene expression during a short burst of terminal differentiation makes them susceptible to the bottleneck effect during RNA processing. This question needs further study to both define the details of this susceptibility as well as develop therapeutic interventions.
Conflict-of-interest disclosure: The authors declare no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal