Abstract
Introduction Diffuse large B-cell lymphoma (DLBCL) is a genetically, phenotypically, and clinically heterogenous disease. Multiple molecular classification systems using either genomic aberrations or gene expression profiles are used to delineate subgroups with shared biology. The cell of origin (COO) gene expression-based classification system stratifies DLBCLs into germinal center B-cell-like (GCB) and activated B-cell-like subgroups of DLBCL. More recently, the double-hit signature (DHITsig) has been used to further stratify GCB DLBCLs into DHITsig+ and DHITsig-, where the former is characterized by a gene expression profile reminiscent of the GC dark zone and inferior outcomes. Currently, patterns of somatic driver mutations can only partially explain the expression of DHITsig in some tumors. Genetic features such as DNA methylation have been shown to be associated with transcriptional repression and altered chromatin structure. We hypothesized that resolving DNA methylation patterns genome-wide using long read whole genome sequencing (LR-WGS) may reveal epigenetic differences associated with differential gene expression profiles within the DHITsig+ and DHITsig- subgroups and in the context of histologic transformation from follicular lymphoma.
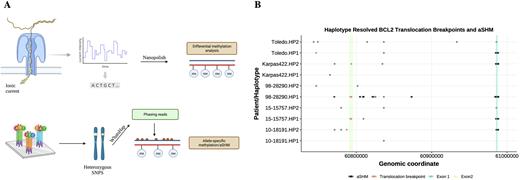
Methods Whole genome sequencing data of 10 patient samples and 4 cell line GCB DLBCL samples were generated using both short read (SR-WGS; Illumina) and LR-WGS (Oxford Nanopore Technologies, PromethION). The complementary approach combines somatic variant calling from highly accurate SR data with DNA methylation calling from LR data. GCB samples were selected based on the NanoString DLBCL90 assay and further subdivided into DHITsig+ (N=4) and DHITsig- (N=3). RNA sequencing data of 9 patient samples and 3 GCB DLBCL cell lines were generated and analyzed using Salmon and DESeq2. Simple somatic mutations from SR-WGS data were called using an ensemble of somatic variant callers, which confirmed all cases to belong to the EZB genetic subgroup. Methylated bases were called with Nanopolish followed by a differential methylation analysis using the DSS package in R to identify differences in methylation between both subgroups. Differentially methylated regions (DMRs) were annotated for GC B-cell specific superenhancers, CpG islands, and genes. Using WhatsHap and Nanomethphase, heterozygous single nucleotide polymorphisms (SNPs) from SR and LR data were used to phase LRs into haplotypes. Haplotype analyses were performed to determine the phase of aberrant somatic hypermutation (aSHM) and methylation patterns in the context of proximal translocation breakpoints.
Results Most DMRs were found to be associated with regulatory regions such as promoters, superenhancers, and CpG islands. We found methylation differences between DHITsig+ and DHITsig- subgroups in 1430 superenhancers known to be active in GC B-cells. Within these superenhancer regions, DHITsig+ tumors were typically hypomethylated relative to DHITsig-. Several DHITsig classifier genes, including WNK2, TNFSF8, and IL21R, had methylation patterns inverted relative to the typical expression pattern (i.e. low methylation with high expression). Similarly, among 633 differentially methylated promoters, several including TCF3 and WNK2 demonstrated the same inverted methylation pattern. CpG islands showed distinct patterns of methylation in both subgroups that were inversely associated with expression, this also included genes differentially expressed between DHITsig+ and DHITsig- tumors. The DHITsig- group had increased levels of methylation of DHITsig genes in both promoters and gene bodies suggesting a functional role for gene body methylation. Finally, we observed that a high density of mutations near the BCL2 transcription start site consistently phased into the same haplotype as the BCL2-IGH translocation in five tumours, demonstrating the allele specificity of aSHM associated with these events (Panel B).
Conclusions These results demonstrate differences in methylation between the subgroups. The association of hypomethylated superenhancers, promoters, and CpG islands with genes over-expressed in DHITsig+ suggests these methylation patterns may contribute to the DHITsig phenotype. These results may aid in improving GCB DLBCL patient classification and reveal factors contributing to its heterogeneity.
Disclosures
Steidl:Abbvie: Consultancy; Bayer: Consultancy; Bristol Myers Squibb: Consultancy; Curis Inc: Consultancy; Epizyme: Research Funding; Roche: Consultancy; Seattle Genetics: Consultancy; Trillium Therapeutics: Research Funding. Scott:Roche: Research Funding; AstraZeneca: Consultancy, Honoraria; Incyte: Consultancy; Janssen: Consultancy, Research Funding; Abbvie: Consultancy; NanoString: Patents & Royalties.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal