Abstract
Introduction Graft versus host disease (GVHD) after allo-HSCT is dependent on conditioning regimen, GVHD prophylaxis, HLA matching, as well as recipient-donor discrepancies in sex, age, and CMV status. GVHD is a leading cause of transplant-related mortality with incidence varying between 30% to 70%. Few machine learning (ML) models have been implemented to accurately quantify a patient's risk for aGVHD development and subsequent severity with mixed results, utilizing various ML methodologies of analyzing clinical data, genetic differences, and biomarkers. Comparing across four different ML algorithms, we demonstrate exceptionally high accuracy for prediction of acute GVHD occurrence, severity (Grade I-II vs III-IV), disease relapse, and survival.
Materials and Methods Data on 868 adult patients (age 18-76) that received either MA or RIC for any transplant indication between January 2004 and July 2018 at The Ohio State University James Cancer Center was analyzed. Predictive models were developed utilizing four different machine learning algorithms: logistic regression, k-nearest neighbors (KNN), decision tree with hyperparameter tuning, and multilayer perceptron (MLP) neural networks. Models were trained and tuned using K-fold cross validation, and prediction performance was quantified by the receiving operating characteristic area-under-the-curve (ROC AUC) and accuracy. Features included patient demographics (age, gender, KPS/ECOG, race, disease type, disease status, donor type, graft type, sex match, HLA match, conditioning regimens groups, use of (ATG), use of alemtuzumab, GVHD prophylaxis type, date of engraftment, date of aGVHD diagnosis, organ involvement of aGVHD, steroid treatment, specific steroid therapy, date of chronic GVHD diagnosis, relapse date, date of death, and cause of death was collected. The dataset was preprocessed through the standardization of numerical features and encoding of categorical features. This dataset was separated pre-analysis into cohorts - 70% exploratory and 30% validation.
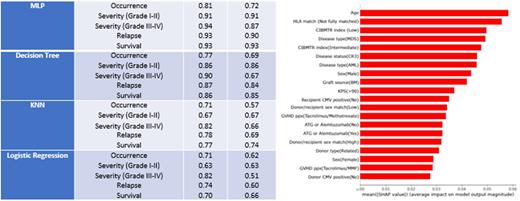
Results All 868 patients were included, verified, and data cleaned. The algorithms predicted with high accuracy - >80% for GVHD occurrence and >90% for Grade I-II and Grade III-IV acute GVHD, relapse, and survival. MLP provided the most accurate predictions when compared to decision tree, KNN and logistic regression algorithms. SHapley Additive exPlanations (SHAP) analysis was used to demonstrate the top 20 features contributing to the overall output. SHAP analysis for acute GVHD occurrence is shown for MLP algorithm with age and HLA match as the most critical features.
Conclusions We demonstrate the results of utilizing advanced ML algorithms for highly accurate prediction of acute GVHD occurrence, severity (Grade I-II vs III-IV), disease relapse, and survival. The difference between the models' performances may be due to the disproportionate sizes of cohorts in Grade III-IV acute GVHD as opposed to relapse and survival analyses. The risk factors identified as being most influential in affecting outcomes by our ML algorithms are consistent with those historically reported. While the importance of these features are similar to prior models, the ability of the models used to generate highly accurate results is novel contribution. The improved accuracy of our methods compared to previous reports may be due to several factors. MLP algorithm with one or two layers have ample hidden neurons to find patterns to achieve superior and robust classification for GVHD. Such deep learning models have not been used in prior classification. Decision trees also provide higher accuracy compared to KNN and regression due to the deterministic nature of the model which automates feature selection. Our single center data may also present a more homogeneous population with decreased variability of practice and GVHD prophylaxis approaches. Further studies to validate these algorithms in a more recent cohort are planned.
Disclosures
Choe:Abbvie: Other: independent endpoint review committee for trial. Ranganathan:Plexxikon Inc: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal