Abstract
Hereditary hematopoietic malignancies (HHMs) are hereditary blood cancer syndromes driven by germline pathogenic/likely pathogenic (P/LP) variants. All HHMs to date have followed Mendelian inheritance patterns. The scientific basis and clinical recognition of HHMs has increased with the rapid uptake of next-generation sequencing (NGS) in research and clinical settings. The accurate diagnosis of an HHM also requires NGS panels that sequence the full spectrum of genes involved in HHMs and sequencing techniques that are capable of detecting the various types of variants that drive HHMs (ie, single nucleotide variants, insertions/deletions, and structural events such as copy number changes). One problem in the HHM field, however, is the quality of HHM diagnostic assays is highly variable. Most HHM assays used in 2020, for example, did not sequence all HHM-related genes.
These omissions increase the risk for false negative test results, which place HHM patients at risk for complications such as donor-derived leukemias (when stem cells from a matched related donor who unknowingly carries an HHM-related variant are provided to an affected recipient), missed opportunities for cancer screening (in HHMs that also carry an increased risk for solid tumors, such as Li Fraumeni syndrome), and reduce opportunities for cascade testing of family members who may benefit from genetic counseling and personalized cancer screening. It is unclear how the landscape of HHM diagnostic testing has changed since the completion of an original analysis of commercial testing practices in 2020. To this end, we queried the US National Center for Biotechnology Information's Genetic Testing Registry to identify all newly offered HHM test offerings since our original analysis. We evaluated all genes on HHM-related panels via a binary matrix approach. We calculated the proportion of panels that sequenced each HHM gene of interest and queried the Online Mendelian Inheritance in Man catalog to confirm the association of each HHM-related gene and a relevant HHM phenotype.
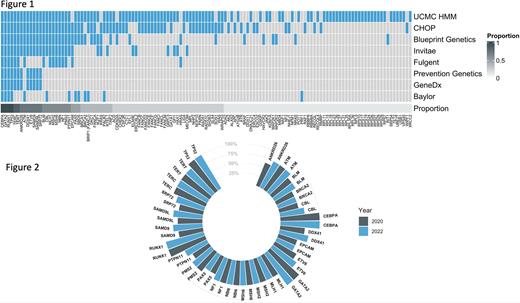
We analyzed HHM panels from eight companies (Fig. 1). Only four genes (CEBPA, GATA2, RUNX1, TP53) were sequenced by all eight panels. An additional 25 genes were sequenced on 50% or more of the panels. Only 25% of HHM panels sequenced CHEK2, the most common HHM variant, and only 75% of HHM panels sequenced DDX41, which was previously considered the gene that is most commonly altered in HHMs. Several important HHM genes, such as BRCA1, BRCA2, CSF3R, FANCA, and MBD4 appeared on only a minority of panels. Furthermore EPCAM, which is not implicated in the etiology or pathogenesis of any known HHM, was present on half of all panels. Trends in analysis of the 25 most-analyzed HHM-related genes from 2020 to January 2022 are shown in Fig. 2. Most HHM assays that previously omitted critical HHM genes made no changes to their panels over this time period. This stagnancy persisted despite multiple associations between candidate genes and HHM phenotypes in public databases. Only one laboratory made significant changes to their panel by adding eight HHM-related genes (ANKRD26, CBL, DDX41, ETV6, PTPN11, SAMD9, SAMD9L, SRP72).
Here, we provide further evidence that the majority of HHM diagnostic assays remain inadequate for the accurate of diagnosis of these syndromes. Stagnation in the quality of HHM diagnostic assays has occurred despite the discovery of novel HHM-related genes, as well as prior publication depicting these inconsistencies. In order to reduce the risk of falsely negative diagnostic reports, the quality of HHM diagnostic testing must be improved so that the most common drivers of HHMs, such as CHEK2 and DDX41, as well as highly penetrant HHMs, such as ETV6, GATA2, and RUNX1, are universally sequenced and analyzed.
Disclosures
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal