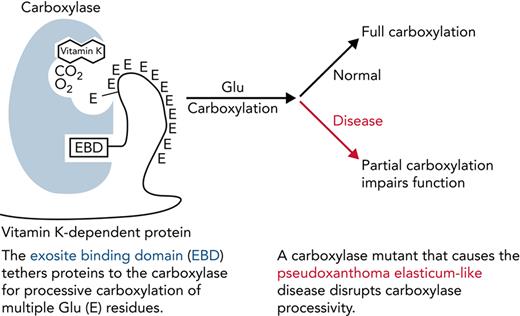
Key Points
Impaired processivity in a carboxylase mutant causes partial carboxylation that explains reduced VKD protein function.
Results suggest that incomplete carboxylation may also occur under other conditions (eg, low vitamin K or during warfarin therapy).
Abstract
γ-Glutamyl carboxylase (GGCX) generates multiple carboxylated Glus (Glas) in vitamin K–dependent (VKD) proteins that are required for their functions. GGCX is processive, remaining bound to VKD proteins throughout multiple Glu carboxylations, and this study reveals the essentiality of processivity to VKD protein function. GGCX mutants (V255M and S300F) whose combined heterozygosity in a patient causes defective clotting and calcification were studied using a novel assay that mimics in vivo carboxylation. Complexes between variant carboxylases and VKD proteins important to hemostasis (factor IX [FIX]) or calcification (matrix Gla protein [MGP]) were reacted in the presence of a challenge VKD protein that could potentially interfere with carboxylation of the VKD protein in the complex. The VKD protein in the complex with wild-type carboxylase was carboxylated before challenge protein carboxylation occurred and became fully carboxylated. In contrast, the V255M mutant carboxylated both forms at the same time and did not completely carboxylate FIX in the complex. S300F carboxylation was poor with both FIX and MGP. Additional studies analyzed FIX- and MGP-derived peptides containing the Gla domain linked to sequences that mediate carboxylase binding. The total amount of carboxylated peptide generated by the V255M mutant was higher than that of wild-type GGCX; however, the individual peptides were partially carboxylated. Analysis of the V255M mutant in FIX HEK293 cells lacking endogenous GGCX revealed poor FIX clotting activity. This study shows that disrupted processivity causes disease and explains the defect in the patient. Kinetic analyses also suggest that disrupted processivity may occur in wild-type carboxylase under some conditions (eg, warfarin therapy or vitamin K deficiency).
Introduction
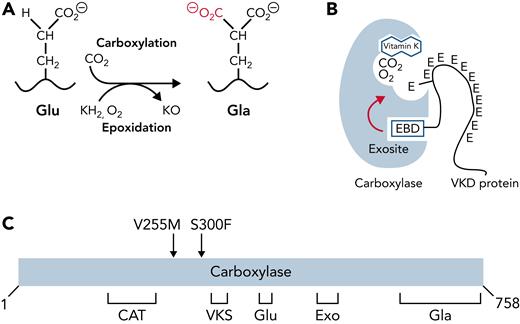
Vitamin K–dependent (VKD) proteins are essential to hemostasis, because several of these proteins function in coagulation and platelet activation.1-3 The major site of synthesis of VKD hemostatic factors is the liver; however, many of these proteins are also expressed in extrahepatic tissues, where they have roles beyond coagulation (eg, inflammation, barrier function, and phagocytosis).4-6 Nonhemostatic VKD proteins have also been identified, which have diverse roles that include regulation of calcification.7 VKD proteins contain carboxylated Glu (Gla) residues required for activity, which are generated by a single γ-glutamyl carboxylase.8 The importance of VKD proteins to human health makes it essential to understand how they become activated by the carboxylase and how disruption of normal VKD protein carboxylation leads to disease.
The carboxylase uses vitamin K epoxidation to drive Glu carboxylation to Gla (Figure 1A). The vitamin K epoxide product must be recycled for continuous carboxylation, which is accomplished by the vitamin K epoxide reductase (VKORC1).9,10 VKORC1 and the carboxylase reside in the endoplasmic reticulum, where VKD proteins are modified during their secretion. VKD proteins are selectively modified because they contain a sequence that mediates high-affinity binding to an exosite in the carboxylase (Figure 1B). In most cases, this sequence (called the EBD) is a propeptide adjacent to the Gla domain, which is cleaved after carboxylation. The mechanism by which the carboxylase modifies VKD proteins is only partly understood. Functional regions important for catalysis and VKD protein interaction have been identified (Figure 1C)11-23; however, carboxylase residues that regulate VKD protein carboxylation are essentially unknown.
Carboxylase function and disruption in PXE-like disease. (A) The carboxylase performs 2 reactions: epoxidation of reduced vitamin K (KH2) to vitamin K epoxide (KO) and carboxylation where CO2 addition to Glu generates carboxylated Glu (Gla). (B) VKD proteins contain an exosite-binding domain (EBD) that mediates high-affinity binding through the carboxylase exosite and also activates Glu catalysis (red arrow). Multiple Glu residues are converted to Gla by a processive mechanism in which VKD proteins remain bound to the carboxylase until the Gla domain is fully carboxylated.27,28 (C) Known functional regions of the carboxylase include those facilitating catalysis (CAT), VKD protein binding (VKS, EXO), Glu binding (Glu), and the carboxylase Gla domain (Gla). Most of the residues whose mutations cause disease reside in regions where the function of the carboxylase is unknown. This study shows that impaired processivity in the V255M mutant results in PXE-like disease.
Carboxylase function and disruption in PXE-like disease. (A) The carboxylase performs 2 reactions: epoxidation of reduced vitamin K (KH2) to vitamin K epoxide (KO) and carboxylation where CO2 addition to Glu generates carboxylated Glu (Gla). (B) VKD proteins contain an exosite-binding domain (EBD) that mediates high-affinity binding through the carboxylase exosite and also activates Glu catalysis (red arrow). Multiple Glu residues are converted to Gla by a processive mechanism in which VKD proteins remain bound to the carboxylase until the Gla domain is fully carboxylated.27,28 (C) Known functional regions of the carboxylase include those facilitating catalysis (CAT), VKD protein binding (VKS, EXO), Glu binding (Glu), and the carboxylase Gla domain (Gla). Most of the residues whose mutations cause disease reside in regions where the function of the carboxylase is unknown. This study shows that impaired processivity in the V255M mutant results in PXE-like disease.
Several regulatory mechanisms affect the efficiency of VKD protein carboxylation. For example, binding of the EBD in VKD proteins to the carboxylase stimulates Glu catalysis (Figure 1B),24 and binding of both the EBD and Glu increases the affinity of the carboxylase for vitamin K.25 The carboxylase is also regulated by substrate-assisted catalysis, where vitamin K epoxidation occurs only in the presence of Glu residues,22,26 which is important in preventing a highly reactive vitamin K intermediate from modifying other molecules when Glu is not present. Finally, carboxylation is regulated through carboxylase processivity, in which VKD proteins remain bound to the carboxylase throughout the multiple Glu-to-Gla conversions.27,28 Processivity is indispensable for the function of other enzymes that perform repetitive reactions (eg, DNA polymerases).29 Other processive enzymes have been extensively studied; however, the processive mechanism for the carboxylase is poorly understood.
Disruption of VKD protein carboxylation causes disease. Naturally occurring mutations in VKORC1 or the carboxylase cause bleeding defects,30-32 and mice lacking either gene die approximately at birth as a result of hemorrhaging.33,34 Some patients with carboxylase mutations also show defects in skin (ie, excessive calcification of elastic fibers in the dermis associated with significant folding and sagging of skin).35-39 The phenotype is due at least in part to defective carboxylation of matrix Gla protein (MGP), an inhibitor of calcification, and is referred to as pseudoxanthoma elasticum (PXE)–like because of its similarity to the PXE disease caused by mutations in a different gene, ABCC6.40 The mechanisms by which carboxylase mutations cause the PXE-like phenotype are not well understood.
We studied the consequences of 2 carboxylase mutants, V255M and S300F, identified in a PXE-like patient with defective clotting and aberrant calcification.36 Both mutations occurred in regions of unknown function in the carboxylase (Figure 1C), and analysis using small peptides derived from VKD proteins did not explain the PXE-like phenotype.36 We therefore developed approaches to study the mutants using more natural VKD substrates that could assess the complex carboxylase reaction. These biochemical assays revealed impaired processivity that resulted in a partially carboxylated VKD product. Cellular analysis showed defective factor IX (FIX) activity. This study linking impaired carboxylase processivity to disease explains the patient phenotype and reveals the essentiality of carboxylase processivity to normal physiology.
Methods
Functional assays
FLAG-tagged V255M and S300F mutants were generated and subcloned into a vector for insect cell expression (supplemental Figure 1) and then expressed in SF21 cells and purified and quantitated as previously reported.41,42 Mutant and wild-type carboxylases were tested in 3 different assays. One assay monitored the overall reaction, which comprises binding of VKD protein to the carboxylase, catalysis of Glus to Glas, and VKD protein release. Carboxylase variants were reacted with FIX- or MGP-derived peptides that contained the EBD and Gla domains (FIX−18/+41 and MGP1-64, respectively; supplemental Table 1). Individual peptides (1 μM) were incubated at 25°C in a reaction cocktail (160 μL) containing 500 mM of NaCl, 50 mM of BES (pH, 6.9), 2.5 mM of dithiothreitol, 0.16% CHAPS, 0.16% of phosphatidyl choline, 200 μM of KH2, and 1.3 mM of [14C]-CO2. Aliquots withdrawn at timed intervals were quenched in sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis (PAGE) loading dye, followed by gel electrophoresis and PhosphorImager analysis. [14C]–bovine serum albumin and a [14C]-peptide standard were analyzed in parallel to quantitate [14C]-CO2 counts, which were converted to pmol of Gla as previously described.20 The [14C]-peptide standard was generated by carboxylation of FIX−18/+41 with wild-type carboxylase, followed by precipitation with chloroform/methanol,17 high-performance liquid chromatography purification, and quantitation by scintillation counting.
A second assay studied the specific Glu-to-Gla catalytic reaction by monitoring the carboxylation of VKD protein–carboxylase complexes. Complexes were generated in SF21 cells coexpressing full-length VKD protein and carboxylase variants as previously described,28 followed by immunopurification. The FIX-carboxylaseFLAG complex was isolated using an antibody against the FIX heavy chain (ESN1; American Diagnostica) immobilized to Sepharose. MGPFLAG-carboxylase complexes were isolated from cells coexpressing MGPFLAG and untagged carboxylase variants, using anti-FLAG agarose (Sigma-Aldrich). MGPFLAG was generated using an MGP complementary DNA (Open Biosystems) and overlap polymerase chain reaction, which added an Ala3-FLAG epitope at the C-terminus. Complexes on the resins were monitored for carboxylation by incubation in the reaction cocktail described above, followed by quenching of timed aliquots with SDS-PAGE loading buffer. After heating at 70°C and centrifugation to pellet resin, the samples were subjected to gel electrophoresis and PhosphorImager analysis. Aliquots were also tested for the amount of complex bound to resin by assaying for epoxidation of reduced vitamin K, as before,20 and by analysis in a western blot using anti-carboxylase antibody,16 after elution from resin in SDS-PAGE loading buffer.
The third assay monitored carboxylase processivity by using a challenge assay in which VKD protein–carboxylase complexes were reacted in the presence of a second VKD substrate.28 FIX-carboxylaseFLAG and MGPFLAG-carboxylase complexes isolated as described above were reacted in the presence of FIX−18/+41 or MGP1-64 peptides, respectively. The reaction cocktail was the same as that described above except for the addition of peptide (1 μM) and less KH2 (10 μM), which was used to mimic the low vitamin K levels that exist in vivo.43 [14C]-CO2 incorporation into challenge VKD substrate and VKD protein in the complexes was quantitated using SDS-PAGE and PhosphorImager analysis.
Results
Analysis of EBD-Gla domain–linked substrate reveals surprisingly higher levels of carboxylation by V255M than by wild-type carboxylase
S300F and V255M mutants identified in a patient with defective clotting and aberrant calcification were initially analyzed in an activation assay. A Glu-containing peptide derived from the Gla domain was coincubated with a separate peptide containing an EBD, which stimulates Glu catalysis24 (Figure 1). When a range of EBD peptide concentrations were tested, half-maximal activity required higher peptide concentrations for the V255M mutant than for wild-type carboxylase (supplemental Figure 1). The results suggested mildly weakened EBD interaction with the V255M carboxylase. S300F carboxylase was essentially inactive.
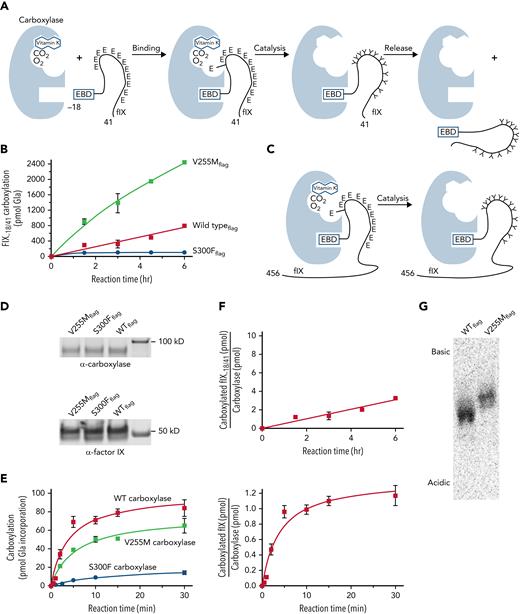
Subsequent analysis used a FIX-derived substrate (ie, FIX−18/+41) containing the EBD covalently linked to the Gla domain (Figure 2A; supplemental Table 1). Linkage of the 2 domains is significant because carboxylase affinity for the Gla domain is orders of magnitude lower than for the EBD; however, linkage of the EBD to the Gla domain results in a high local concentration of Glu residues for efficient carboxylation. FIX−18/+41 was incubated in reactions containing [14C]-CO2, KH2, and equivalent amounts of purified carboxylase variants. Aliquots withdrawn at timed intervals were quenched by SDS-PAGE loading dye, followed by gel electrophoresis and PhosphorImager analysis. [14C]-CO2 incorporation into the peptides was quantitated by comparison with [14C]-standards, and [14C]-CO2 counts were converted to pmol of Gla as previously described.20 The results revealed surprisingly higher levels of carboxylated FIX−18/+41 generated by V255M than by wild-type carboxylase (400%; Figure 2B). In contrast, S300F activity was low (ie, ∼10% that of wild-type carboxylase).
V255M carboxylase generates higher levels of modified FIX than wild-type carboxylase. (A) Carboxylation of Glu (E) to Gla (Y) residues in FIX occurs in 3 steps (ie, binding, catalysis, and release). The overall reaction was studied using FIX−18/+41, which contains the 18–amino acid EBD covalently linked to the 41–amino acid Gla domain with 12 Glu residues. The EBD in FIX is a propeptide that is cleaved subsequent to carboxylation. (B) FLAG-tagged wild-type (WT) and mutant carboxylases were immunopurified using anti-FLAG antibody and quantitated in western blots with anti-FLAG and anti-carboxylase antibodies. Equivalent amounts of variant carboxylases were then reacted with FIX−18/+41, followed by SDS-PAGE and PhosphorImager analysis. (C-E) The specific catalytic step was monitored using FIX-carboxylase complexes generated in cells expressing FIX and individual variant carboxylases. After isolation on anti-FIX resin, aliquots were monitored for the amount of FIX-carboxylase complex by an epoxidation assay,20 which was confirmed by western blot analysis. Equivalent amounts of complex (6 pmol) were then monitored for pmol of Gla incorporation into FIX using gel electrophoresis and PhosphorImager analysis. (F) The amounts of carboxylated FIX−18/+41 or FIX in the complex in panels B and E were compared with the amount of WT carboxylaseFLAG to determine the times to complete the reactions. The time for an individual FIX-carboxylase complex to become fully carboxylated was ∼15 minutes (lower panel), whereas FIX−18/+41 was carboxylated in ∼75 minutes (as indicated by a ratio of 1 for FIX−18/+41/carboxylase levels in the upper panel). (G) FIX−18/+41 was reacted with WT or mutant carboxylases as in panel B, and the products were precipitated with chloroform/methanol17 and resuspended in 25 mM of ammonium bicarbonate. Aliquots were quantitated by scintillation counting, and similar levels of counts were subjected to isoelectric focusing using a pH 3-10 Criterion IEF gel (Bio-Rad Laboratories) and the buffers recommended by the manufacturer. This analysis revealed a more basic FIX−18/+41 product with the V255M mutant than with WT carboxylase, indicating partial carboxylation.
V255M carboxylase generates higher levels of modified FIX than wild-type carboxylase. (A) Carboxylation of Glu (E) to Gla (Y) residues in FIX occurs in 3 steps (ie, binding, catalysis, and release). The overall reaction was studied using FIX−18/+41, which contains the 18–amino acid EBD covalently linked to the 41–amino acid Gla domain with 12 Glu residues. The EBD in FIX is a propeptide that is cleaved subsequent to carboxylation. (B) FLAG-tagged wild-type (WT) and mutant carboxylases were immunopurified using anti-FLAG antibody and quantitated in western blots with anti-FLAG and anti-carboxylase antibodies. Equivalent amounts of variant carboxylases were then reacted with FIX−18/+41, followed by SDS-PAGE and PhosphorImager analysis. (C-E) The specific catalytic step was monitored using FIX-carboxylase complexes generated in cells expressing FIX and individual variant carboxylases. After isolation on anti-FIX resin, aliquots were monitored for the amount of FIX-carboxylase complex by an epoxidation assay,20 which was confirmed by western blot analysis. Equivalent amounts of complex (6 pmol) were then monitored for pmol of Gla incorporation into FIX using gel electrophoresis and PhosphorImager analysis. (F) The amounts of carboxylated FIX−18/+41 or FIX in the complex in panels B and E were compared with the amount of WT carboxylaseFLAG to determine the times to complete the reactions. The time for an individual FIX-carboxylase complex to become fully carboxylated was ∼15 minutes (lower panel), whereas FIX−18/+41 was carboxylated in ∼75 minutes (as indicated by a ratio of 1 for FIX−18/+41/carboxylase levels in the upper panel). (G) FIX−18/+41 was reacted with WT or mutant carboxylases as in panel B, and the products were precipitated with chloroform/methanol17 and resuspended in 25 mM of ammonium bicarbonate. Aliquots were quantitated by scintillation counting, and similar levels of counts were subjected to isoelectric focusing using a pH 3-10 Criterion IEF gel (Bio-Rad Laboratories) and the buffers recommended by the manufacturer. This analysis revealed a more basic FIX−18/+41 product with the V255M mutant than with WT carboxylase, indicating partial carboxylation.
The assay shown in Figure 2B monitored multiple steps in carboxylation (ie, binding, catalysis, and release; Figure 2A), and further analysis specifically monitored catalysis (Figure 2C-E). This assay was possible because FIX remains bound to wild-type carboxylase long after it becomes fully carboxylated.44 Preformed complexes between carboxylase variants and full-length FIX were generated in cells, which were cultured in the absence of vitamin K so that FIX was uncarboxylated. The complexes were isolated on anti-FIX antibody resin to remove free carboxylase that would have interfered with analysis, using anti-FIX antibody that reacts to a region of FIX several hundred amino acids away from the Gla domain. Some FIX not in a complex with the carboxylase was also captured on the resin, which control experiments showed did not interfere with the assay (supplemental Figure 2). Specifically, the uncomplexed FIX was not carboxylated, presumably because its concentration was far below the Km for FIX binding by the carboxylase and because immobilization prevented interaction with carboxylase in the FIX-carboxylase complexes.
To monitor carboxylation, equivalent amounts of complex containing mutant or wild-type carboxylase and FIX were incubated in reactions containing KH2 and [14C]-CO2, and timed aliquots quenched with SDS gel loading buffer were subjected to gel electrophoresis and PhosphorImager analysis to monitor [14C]-CO2 incorporation into FIX. [14C]-standards analyzed in parallel allowed the conversion of [14C]-CO2 counts to pmol of Gla, as previously described.20 The values were compared with the amount of complex as quantitated in anti-carboxylase western blots (Figure 2D) and by an epoxidase assay that gave similar carboxylase levels. The results indicated full carboxylation of FIX by wild-type carboxylase, consistent with previous studies.28 Gla production by the V255M and S300F mutants was lower than with wild-type carboxylase (Figure 2E). Higher V255M activity in the overall reaction (Figure 2B) was therefore not due to the catalytic step.
A comparison of the rates of catalysis vs the overall reaction was of interest with regard to the V255M carboxylase defect. Wild-type carboxylase catalysis was complete in ∼15 minutes (Figure 2E-F), and this rate for FIX-carboxylase complexes immobilized on resin was similar to that observed with free complex (supplemental Figure 3). The overall reaction for FIX−18/+41 carboxylation took 75 minutes (Figure 2F), similar to the value previously determined for full-length propeptide-containing FIX.28 FIX catalysis was therefore ∼5 times faster than the overall reaction with wild-type carboxylase. In contrast, the overall reaction with V255M carboxylase was faster than that with wild-type carboxylase, whereas catalysis was slower (Figure 2B,E). These differences raised the question of whether the V255 mutant fully carboxylates FIX.
Isoelectric focusing was performed to assess the FIX−18/+41 carboxylation products. This approach separates proteins based on charge, and the conversion of Glus to Glas adds negative charge to VKD proteins. FIX−18/+41 was carboxylated and precipitated to remove [14C]-CO2, and an aliquot of resuspended sample was subjected to scintillation counting. The S300F sample did not show detectable carboxylation and was not processed any further. Isoelectric focusing revealed a more basic product generated by V255M than by wild-type carboxylase (Figure 2G), indicating undercarboxylation. Therefore, although the total amount of Gla generated by the V255M mutant was higher than with wild-type carboxylase (Figure 2B), the individual peptides were not completely carboxylated (Figure 2G). The results suggested impaired processivity in the V255M mutant, which was subsequently tested.
Processive FIX carboxylation is impaired in the V255M mutant
Carboxylase processivity was tested using a challenge assay in which carboxylase–VKD protein complexes were reacted in the presence of excess challenge VKD substrate, and carboxylation of both VKD forms was monitored (Figure 3A). Our previous development of this assay showed that wild-type carboxylase is processive, resulting in full carboxylation of FIX in the complex in the presence of the challenge VKD protein.28 The challenge substrate used in the current study was FIX−18/+41, which has kinetic parameters (ie, Km and kcat) similar to propeptide-containing full-length FIX28 and therefore could potentially compete with FIX in the complex for carboxylation.
Processive carboxylation of FIX is impaired in the V255M mutant. (A) A complex between the carboxylase and FIX was reacted in the presence of a challenge FIX (FIX−18/+41), and carboxylation of both FIX forms was monitored. (B-E) Independent gels were used to monitor FIX−18/+41 and fIX in the wild-type (WT) or V255M complexes because of differences in their size, and [14C]-standards were included on the gels to allow quantitation of carboxylation products. (F-G) FIX−18/+41 was carboxylated after FIX in the complex with WT carboxylase but occurred simultaneously with FIX in the complex with V255M carboxylase. FIX carboxylation was much lower in the complex with the V255M mutant than with WT carboxylase. It was also lower than observed in the reaction monitoring catalysis (Figure 2E), which may have been due to the lower KH2 concentrations used in the processivity assay. (H) S300F carboxylation of both FIX in the complex and FIX−18/+41 was poor.
Processive carboxylation of FIX is impaired in the V255M mutant. (A) A complex between the carboxylase and FIX was reacted in the presence of a challenge FIX (FIX−18/+41), and carboxylation of both FIX forms was monitored. (B-E) Independent gels were used to monitor FIX−18/+41 and fIX in the wild-type (WT) or V255M complexes because of differences in their size, and [14C]-standards were included on the gels to allow quantitation of carboxylation products. (F-G) FIX−18/+41 was carboxylated after FIX in the complex with WT carboxylase but occurred simultaneously with FIX in the complex with V255M carboxylase. FIX carboxylation was much lower in the complex with the V255M mutant than with WT carboxylase. It was also lower than observed in the reaction monitoring catalysis (Figure 2E), which may have been due to the lower KH2 concentrations used in the processivity assay. (H) S300F carboxylation of both FIX in the complex and FIX−18/+41 was poor.
Preformed FIX-carboxylase complexes generated in cells were isolated on anti-FIX antibody resin to remove free carboxylase. The anti-FIX antibody reacts to a region distinct from the FIX−18/+41 sequences and did not cross-react with the FIX−18/+41 peptide (data not shown). Equivalent amounts of FIX-carboxylase complexes were incubated in reactions containing [14C]-CO2 along with FIX−18/+41 at a concentration (1 μM) 50-fold in excess over the complex (0.02 μM). The FIX−18/+41 peptide concentration was 100-fold higher than that of the uncomplexed FIX also captured on the anti-FIX resin, which as described in the previous section, was not carboxylated. Carboxylation was initiated by the addition of KH2, and timed aliquots were quenched with SDS loading buffer and subjected to gel electrophoresis and PhosphorImager analysis. [14C]-standards were processed in parallel to quantitate [14C]-CO2 incorporation into FIX in the complex and FIX−18/+41, which was converted to pmol of Gla as previously described.20 FIX in the complex with wild-type carboxylase showed a mobility shift as the 12 Glus underwent conversion to Glas (Figure 3B). [14C]-CO2 incorporation into the discrete slower-migrating FIX form observed at the end of the reaction was compared with the amount of complex determined by an anti-carboxylase western blot and an epoxidase assay, which indicated full FIX carboxylation, similar to previous results.44 Carboxylation of FIX−18/+41 occurred subsequent to that of FIX in the complex (Figure 3B,C,F). Some carboxylated FIX−18/+41 was observed before the carboxylation of FIX in the complex was complete, possibly because of nonsynchronous carboxylation of individual complexes. A control experiment analyzing carboxylation with free carboxylase showed simultaneous carboxylation of both proteins (supplemental Figure 4), indicating that the delay in FIX−18/+41 carboxylation was specific to the FIX-carboxylase complex.
The results with V255M-FIX complexes were strikingly different. FIX−18/+41 carboxylation occurred at the same time as that of FIX in the complex (Figure 3D-E,G). Carboxylation of FIX in the V255M-FIX complex was eightfold lower than that observed with wild-type carboxylase (Figure 3F-G), and the heterogeneous mixture of carboxylated FIX forms did not transition into the slower-migrating form observed with wild-type carboxylase (Figure 3B,D). A similar pattern of FIX carboxylation was observed even in the absence of FIX−18/+41 (supplemental Figure 5). Decreased V255M carboxylation of FIX was not due to inactivation, as indicated by FIX−18/+41 carboxylation (Figure 3G) and an epoxidase assay (data not shown). By the end of the reaction, the amount of carboxylated challenge protein vs FIX in the complex was significantly higher with the V255M mutant than wild-type carboxylase. The results indicate impaired processivity in the V255M carboxylase.
Carboxylation of FIX in the S300F-FIX complex was barely detectable (Figure 3H). Carboxylation was lower than that observed in the assay for catalysis (Figure 2E), possibly because of the lower vitamin K concentration used in the processivity assay. FIX−18/+41 carboxylation by the S300F mutant was also poor, and low levels precluded quantitation.
Cellular V255M carboxylation results in decreased FIX clotting activity
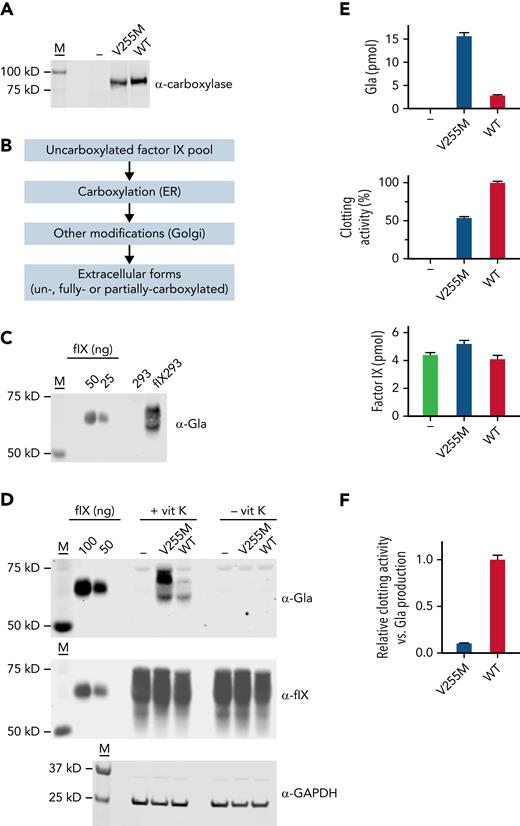
Complementary DNAs encoding V255M and wild-type carboxylases were subcloned into pCMV6-AC (Origene) and then stably expressed in FIX 293 cells (Figure 4A), which were previously edited to eliminate endogenous carboxylase.18 To assess multiple FIX turnovers (ie, the overall reaction) by the carboxylase as in the biochemical studies, a pool of uncarboxylated FIX was generated in cells lacking vitamin K, and secreted FIX was analyzed after exchanging the cells into media containing vitamin K (Figure 4B). Serum-free media was used throughout so that bovine serum VKD proteins did not interfere with analysis. Carboxylation of secreted FIX was monitored in a western blot using a pan-specific anti-Gla antibody generated against a consensus sequence in VKD proteins.45 Antibody reactivity was observed in FIX 293 cells but not in progenitor 293 cells (Figure 4C), which allowed the specific analysis of FIX carboxylation. FIX secreted from the FIX 293 cells migrated differently from the purified plasma control (Figure 4C-D) because of differences in posttranslational modifications, which do not affect activity.46,47
V255M carboxylation in cells generates FIX with decreased clotting activity. (A) FLAG-tagged wild-type (WT) and V255M carboxylases were individually expressed in FIX 293 cells edited to eliminate endogenous carboxylase (−). M indicates molecular weight markers. (B) Cells cultured in the absence of vitamin K resulted in an uncarboxylated intracellular pool of FIX in ∼50-fold excess over the carboxylase, as determined by western blot analysis with anti-FIX and anti-FLAG antibodies and FIX and FLAG standards. Cells were then exchanged into serum-free media containing vitamin K (5 ng/ml) and harvested after 18 hours. During secretion, FIX was carboxylated in the endoplasmic reticulum (ER), and additional modifications occurred in the Golgi (eg, propeptide processing, sulfation, and aspartyl β-hydroxylation). Carboxylation was not obligatory for secretion, and secreted FIX could therefore be a mixture of forms with different degrees of carboxylation. (C) A panspecific anti-Gla antibody45 detected VKD protein in cell-spent media from FIX 293 cells but not the progenitor 293 cells, allowing the carboxylation of FIX expressed in 293 cells to be specifically analyzed. The FIX control was purified human plasma FIX (Enzyme Research Laboratories). (D) Media from cells cultured in the presence or absence of vitamin K (vit K) were analyzed in western blots using antibody against Gla45 or anti-FIX antibody that detects both carboxylated and uncarboxylated FIX.18 Lysates analyzed with anti–glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody indicated similar amounts of cellular material. (E) Media was quantitated for Gla and FIX content by comparison with purified plasma FIX (shown in panel D). Aliquots from the same media samples analyzed in the western blots were assayed for FIX activity in a clotting assay, as previously described.46,67 (F) Clotting activity and Gla content were compared and normalized to a ratio of 1 for FIX secreted from 293 cells expressing WT carboxylase. The ratio of clotting activity to Gla content revealed defective clotting in FIX carboxylated by the V255M mutant.
V255M carboxylation in cells generates FIX with decreased clotting activity. (A) FLAG-tagged wild-type (WT) and V255M carboxylases were individually expressed in FIX 293 cells edited to eliminate endogenous carboxylase (−). M indicates molecular weight markers. (B) Cells cultured in the absence of vitamin K resulted in an uncarboxylated intracellular pool of FIX in ∼50-fold excess over the carboxylase, as determined by western blot analysis with anti-FIX and anti-FLAG antibodies and FIX and FLAG standards. Cells were then exchanged into serum-free media containing vitamin K (5 ng/ml) and harvested after 18 hours. During secretion, FIX was carboxylated in the endoplasmic reticulum (ER), and additional modifications occurred in the Golgi (eg, propeptide processing, sulfation, and aspartyl β-hydroxylation). Carboxylation was not obligatory for secretion, and secreted FIX could therefore be a mixture of forms with different degrees of carboxylation. (C) A panspecific anti-Gla antibody45 detected VKD protein in cell-spent media from FIX 293 cells but not the progenitor 293 cells, allowing the carboxylation of FIX expressed in 293 cells to be specifically analyzed. The FIX control was purified human plasma FIX (Enzyme Research Laboratories). (D) Media from cells cultured in the presence or absence of vitamin K (vit K) were analyzed in western blots using antibody against Gla45 or anti-FIX antibody that detects both carboxylated and uncarboxylated FIX.18 Lysates analyzed with anti–glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody indicated similar amounts of cellular material. (E) Media was quantitated for Gla and FIX content by comparison with purified plasma FIX (shown in panel D). Aliquots from the same media samples analyzed in the western blots were assayed for FIX activity in a clotting assay, as previously described.46,67 (F) Clotting activity and Gla content were compared and normalized to a ratio of 1 for FIX secreted from 293 cells expressing WT carboxylase. The ratio of clotting activity to Gla content revealed defective clotting in FIX carboxylated by the V255M mutant.
The anti-Gla signal was fivefold higher in V255M 293 cells vs cells expressing wild-type carboxylase (Figure 4D-E), consistent with the higher levels of modified FIX observed in the biochemical assay (Figure 2B). Media was also analyzed using an anti-FIX antibody that detects both carboxylated and uncarboxylated FIX.18 FIX was secreted from cells lacking vitamin K or the carboxylase (Figure 4D), indicating that FIX carboxylation was not obligatory for secretion and that FIX in cells containing vitamin K can therefore be a mixture of carboxylated and uncarboxylated forms. The anti-Gla and anti-FIX signals were converted to pmol using FIX standards and adjusted for 12 Gla residues in FIX (Figure 4E), which indicated that only some of the FIX pool (6%) underwent carboxylation. Consequently, the increase in anti-FIX signal in V255M- vs wild-type carboxylase cells was not as dramatic as observed with the anti-Gla signal.
FIX activity was determined by monitoring the ability of samples to restore clotting in FIX-deficient serum. The same media samples analyzed in the western blots were tested for activity, which revealed twofold lower clotting activity for FIX carboxylated by the V255M mutant than by wild-type carboxylase (Figure 4E). A comparison of the amount of clotting activity vs Gla in secreted FIX indicated a 10-fold lower level in cells expressing V255M carboxylase (Figure 4F). The V255M mutant therefore generates defective FIX, consistent with decreased FIX activity in the patient.36 Analysis of carboxylation by isoelectric focusing as in the biochemical studies was not possible, because FIX undergoes other modifications that affect charge (eg, sulfation and Asp β-hydroxylation).
Processive MGP carboxylation by V255M and S300F carboxylases is impaired
The patient with the V255M and S300F mutations shows aberrant calcification, and MGP, which regulates calcification, was therefore also studied. Processivity was tested in the challenge assay using complexes between MGPFLAG and variant carboxylases and a challenge substrate (MGP1-64) that contains the EBD and Gla domain (Figure 5A; supplemental Table 1). The organization of the EBD and Gla domains in MGP is distinct from that in most VKD proteins, having an EBD that is part of the mature protein. MGPFLAG-carboxylase complexes were separated from free carboxylase by isolation on anti-FLAG resin, and equivalent amounts of complexes containing variant carboxylases were then incubated with [14C]-CO2 along with MGP1-64 that was in 50-fold excess over the complexes. After the addition of KH2, timed aliquots were subjected to gel electrophoreses and PhosphorImager analysis (Figure 5B-D), along with a [14C]-standard that allowed [14C]-CO2 quantitation and conversion to pmol of Gla, as before.20 MGPFLAG in the complex with wild-type carboxylase was carboxylated before MGP1-64, whereas V255M carboxylase modified both MGP forms at the same time (Figure 5E-F). The V255M mutant also differed from wild-type carboxylase in producing substantially more carboxylated MGP1-64 than MGPFLAG in the complex (Figure 5C,F). The results indicated impaired processivity. MGPFLAG carboxylation by the S300F mutant was poor but produced significant amounts of carboxylated MGP1-64 (Figure 5D,G).
Processive MGP carboxylation is impaired in the V255M and S300F mutants. (A) Complexes between MGPFLAG and variant carboxylases were reacted in the presence of a challenge peptide (MGP1-64) that contained both the EBD and the entire Gla domain. (B-D) MGPFLAG-carboxylase complexes were isolated on anti-FLAG resin, and the amount of complex was determined by an epoxidase assay, as before.20 Equivalent amounts of complex were then analyzed in the challenge assay. Carboxylation of MGPFLAG in the complexes and MGP1-64 was quantitated by monitoring [14C]-CO2 incorporation into each form, using gel electrophoresis and PhosphorImager analysis. (E-G) MGP1-64 carboxylation occurred after MGPFLAG in the complex with wild-type carboxylase. Both MGP forms were carboxylated at the same time by mutant carboxylases, which produced higher levels of MGP1-64 than MGPFLAG in the complex.
Processive MGP carboxylation is impaired in the V255M and S300F mutants. (A) Complexes between MGPFLAG and variant carboxylases were reacted in the presence of a challenge peptide (MGP1-64) that contained both the EBD and the entire Gla domain. (B-D) MGPFLAG-carboxylase complexes were isolated on anti-FLAG resin, and the amount of complex was determined by an epoxidase assay, as before.20 Equivalent amounts of complex were then analyzed in the challenge assay. Carboxylation of MGPFLAG in the complexes and MGP1-64 was quantitated by monitoring [14C]-CO2 incorporation into each form, using gel electrophoresis and PhosphorImager analysis. (E-G) MGP1-64 carboxylation occurred after MGPFLAG in the complex with wild-type carboxylase. Both MGP forms were carboxylated at the same time by mutant carboxylases, which produced higher levels of MGP1-64 than MGPFLAG in the complex.
MGP carboxylation was also tested in the assay that monitored the overall reaction (Figure 6A), using the MGP1-64 peptide. Purified carboxylase variants were incubated with peptide in reactions containing KH2 and [14C]-CO2, and timed aliquots were subjected to gel electrophoresis and PhosphorImager analysis. A [14C]-standard was included to quantitate [14C]-CO2 incorporation into the peptide and consequent pmol of Gla. MGP1-64 reacted with wild-type carboxylase at a rate similar to that of FIX−18/+41 despite their having different EBD-Gla domain organizations (Figure 6B). V255M carboxylation of MGP1-64 was higher than that of wild-type carboxylase (Figure 6C), similar to the results with FIX−18/+41 (Figure 2B), whereas S300F turnover was poor. MGP catalysis was also analyzed (Figure 6D-F), revealing slower MGPFLAG catalysis by the V255M mutant than by wild-type carboxylase (Figure 6F). A comparison of the rates for the overall reaction vs catalysis showed that the time for wild-type carboxylase to modify the MGP Gla domain (10 minutes) was sixfold faster than the time for the overall reaction (60 minutes). A faster overall reaction and slower catalysis with V255M carboxylase suggest the potential for undercarboxylation of MGPFLAG. Several attempts were made to analyze MGP carboxylation by isoelectric focusing, which were unsuccessful because of solubility issues (as observed by others48-51).
The V255M mutant shows higher levels of modified MGP than wild-type (WT) carboxylase. (A) The overall reaction for MGP carboxylation comprises binding, catalysis, and release, which produces MGP containing 5 Glas (indicated by Ys). (B) MGP1-64 and FIX−18/+41 were incubated with the same amount of WT carboxylase, followed by SDS-PAGE and PhosphorImager analysis to quantitate [14C]-CO2 incorporation into the peptides. (C) The same assay was used to measure MGP1-64 carboxylation by WT and mutant carboxylases, using equivalent amounts of enzyme as determined by western blot analysis with an anti-carboxylase antibody. Higher carboxylase concentrations were used than in panel B, and the duplicate samples that were assayed gave almost identical results and consequent small error bars. The overall reaction for WT carboxylase took 60 minutes, as determined by comparing pmol of Gla in MGP1-64 with the amount of carboxylase. (D-F) The specific catalytic step was monitored using preformed MGPFLAG-carboxylase complexes generated in cells, which were adsorbed to anti-FLAG resin to remove free carboxylase. The amount of carboxylase was quantitated by an epoxidase assay,20 and equivalent amounts of complex were then incubated in reactions containing KH2 and [14C]-CO2, followed by gel electrophoresis and PhosphorImager analysis to monitor [14C]-CO2 incorporation into MGPFLAG.
The V255M mutant shows higher levels of modified MGP than wild-type (WT) carboxylase. (A) The overall reaction for MGP carboxylation comprises binding, catalysis, and release, which produces MGP containing 5 Glas (indicated by Ys). (B) MGP1-64 and FIX−18/+41 were incubated with the same amount of WT carboxylase, followed by SDS-PAGE and PhosphorImager analysis to quantitate [14C]-CO2 incorporation into the peptides. (C) The same assay was used to measure MGP1-64 carboxylation by WT and mutant carboxylases, using equivalent amounts of enzyme as determined by western blot analysis with an anti-carboxylase antibody. Higher carboxylase concentrations were used than in panel B, and the duplicate samples that were assayed gave almost identical results and consequent small error bars. The overall reaction for WT carboxylase took 60 minutes, as determined by comparing pmol of Gla in MGP1-64 with the amount of carboxylase. (D-F) The specific catalytic step was monitored using preformed MGPFLAG-carboxylase complexes generated in cells, which were adsorbed to anti-FLAG resin to remove free carboxylase. The amount of carboxylase was quantitated by an epoxidase assay,20 and equivalent amounts of complex were then incubated in reactions containing KH2 and [14C]-CO2, followed by gel electrophoresis and PhosphorImager analysis to monitor [14C]-CO2 incorporation into MGPFLAG.
Discussion
This study reveals the importance of processive carboxylation in generating the multiple Gla residues in VKD proteins that form a calcium-binding module required for activity. Processivity was studied with V255M and S300F carboxylase mutants present in a patient with defective clotting and aberrant calcification.36 VKD proteins important to hemostasis (FIX) or calcification (MGP) were investigated using a novel challenge assay in which a VKD protein–carboxylase complex was reacted in the presence of exogenous VKD substrate that was in large excess over the complex. FIX and MGP in a complex with wild-type carboxylase were carboxylated before the challenge substrates and became fully carboxylated (Figures 3 and 5). S300F activity was poor with MGP and barely detectable with FIX. In the case of the V255M mutant, FIX and MGP in the complexes were carboxylated at the same time as the challenge protein, the challenge VKD protein was carboxylated at higher levels than the VKD protein in the complex, and FIX carboxylation in the complex was poor. Wild-type and V255M carboxylases also showed significant differences in the relative rates of catalysis vs the overall reaction. Processive complete carboxylation results from the Gla domain becoming fully carboxylated before VKD proteins are released from the carboxylase (Figure 7A). Wild-type carboxylase completed carboxylation of the FIX and MGP Gla domains at rates five- and sixfold faster than the overall reaction, respectively. In contrast, the 2 rates were similar for the V255M mutant, which generated partially carboxylated FIX (Figure 2). When V255M carboxylase was expressed in FIX 293 cells edited by CRISPR/Cas9 to eliminate endogenous carboxylase, FIX with defective clotting activity was produced (Figure 4), as in the patient.36 Impaired processivity can therefore explain the phenotype of the patient.
Carboxylase processivity and disruption in disease. (A) Processivity depends upon the relative rates of catalysis and release. Wild-type (WT) carboxylase completed carboxylation of the FIX Gla domain at a rate fivefold faster than that of the overall reaction. The rates of catalysis and the overall reaction were similar with the V255M mutant, which generated partially carboxylated FIX. (B) WT carboxylase shields a VKD protein undergoing carboxylation from external VKD proteins, which allows full carboxylation. Shielding implicates a transition after Glu carboxylation to an open conformation that allows exit of the carboxylated VKD protein and entrance of an uncarboxylated substrate. A conformational change in the V255M mutant may disrupt this normal process to result in partially carboxylated VKD proteins. Alternatively, weakened interaction between the V255M mutant and VKD proteins could lead to premature release, accounting for partial carboxylation.
Carboxylase processivity and disruption in disease. (A) Processivity depends upon the relative rates of catalysis and release. Wild-type (WT) carboxylase completed carboxylation of the FIX Gla domain at a rate fivefold faster than that of the overall reaction. The rates of catalysis and the overall reaction were similar with the V255M mutant, which generated partially carboxylated FIX. (B) WT carboxylase shields a VKD protein undergoing carboxylation from external VKD proteins, which allows full carboxylation. Shielding implicates a transition after Glu carboxylation to an open conformation that allows exit of the carboxylated VKD protein and entrance of an uncarboxylated substrate. A conformational change in the V255M mutant may disrupt this normal process to result in partially carboxylated VKD proteins. Alternatively, weakened interaction between the V255M mutant and VKD proteins could lead to premature release, accounting for partial carboxylation.
Multiple defects could explain partial VKD protein carboxylation by the V255M mutant. The affinity of the V255M mutant for the EBD of VKD proteins is weaker than that of wild-type carboxylase (supplemental Figure 1), and VKD proteins may not remain bound to the mutant for a sufficient time to achieve full carboxylation. Alternatively, a conformational change in the V255M mutant could affect the normal process by which carboxylated proteins are released. During wild-type carboxylation, VKD proteins are maintained in a closed carboxylase conformation that blocks the access of exogenous substrate and allows the carboxylation of multiple Glu residues (Figure 7B). The model is consistent with the delay in carboxylation of challenge peptide reacted with complexes between VKD proteins and wild-type carboxylase (Figures 3 and 5), which was not observed with free carboxylase (supplemental Figure 4). The model is also supported by previous studies showing that even a small Glu peptide derived from the Gla domain is not carboxylated until after carboxylation of FIX in an FIX-carboxylase complex.28 The results implicate a transition at the end of the catalytic reaction from a closed to an open carboxylase conformation, which allows the exit of carboxylated VKD protein and entrance of new uncarboxylated protein (Figure 7B). Carboxylation alone is not sufficient for the release of VKD proteins, because we found that fully carboxylated FIX either isolated from mammalian cells or generated in vitro remained bound to the carboxylase.44 Interestingly, the presence of exogenous VKD protein accelerated release of carboxylated VKD protein, indicating a release mechanism more complicated than previously appreciated. The V255M mutant, then, may disrupt the normal process of release.
The observation that wild-type carboxylase only shows a small difference in the time to fully carboxylate the Gla domain vs the length of time the carboxylase remains bound to VKD proteins is significant because it suggests that wild-type carboxylase may sometimes generate partially carboxylated VKD proteins. The overall reaction was only five- to sixfold slower than full carboxylation of the Gla domain (Figures 2 and 6); however, the relative rates may vary in vivo. For example, vitamin K levels that control the catalytic rate are low in vivo and vary significantly with diet,52 which could affect processivity. Warfarin therapy that targets VKORC1 to decrease reduced vitamin K levels could also disrupt processivity, explaining previous studies suggesting undercarboxylation with warfarin treatment.53,54 Partially carboxylated VKD forms may therefore be present more frequently than previously appreciated. We note that previous studies reported a much larger difference between the rates of Glu catalysis and release (3000-fold),55,56 which should result in full carboxylation. However, the 3000-fold value was obtained studying small unlinked peptides, comparing the release of an EBD peptide from the carboxylase with catalysis of a separate peptide containing a few Glus. Our results with more natural VKD substrates provide a substantially revised view of carboxylase processivity and the consequent extent of VKD protein carboxylation in vivo.
This study illustrates the value of using both biochemical and cellular approaches to understand how carboxylase mutations cause disease. Biochemical studies are important because some analyses are not possible in cells (eg, studying processivity). In addition, biochemical analyses are direct, whereas the cellular approach that analyzes the carboxylation status of secreted VKD proteins is indirect and affected by the secretory process (eg, quality control mechanisms that eliminate poorly carboxylated VKD proteins).57-59 Although biochemical analyses have a potential limitation (ie, the high salt conditions used in the assays as required for activity), the results predicted consequences consistent with those observed in the cellular studies. The value of biochemical approaches is underscored by 2 recent cellular studies on carboxylase mutations that included V255M.60,61 Both studies assessed carboxylation using antibodies that did not reveal partial carboxylation and indicated wild-type levels of carboxylation by V255M. One of the studies also included a functional analysis, which showed that FVII modified by the mutant had activity (85%) within the normal range.60 We observed decreased FIX clotting activity in cells expressing V255M carboxylase, similar to that in the patient,36 whose phenotype can be explained by biochemical analyses that revealed impaired processivity. Combined biochemical and cellular approaches are therefore highly informative for determining how carboxylase mutations cause disease.
The V255M mutant generated FIX that retained some clotting activity (Figure 4E), and future studies that define the type of partially carboxylated FIX generated by V255M carboxylase will be of interest. The number of Gla residues required for VKD protein function is unknown. In addition, some Glas are likely to be more important than others, as indicated by previous mutagenesis studies where Glu-to-Asp substitutions in the Gla domains of protein C, FX, and prothrombin showed that individual Gla residues contribute differently to function.62-65 A second area of interest will be to determine whether processive carboxylation is the same for all VKD proteins. Widely different affinities have been reported for the EBDs that mediate VKD protein binding to the carboxylase,66 which could affect the length of time a VKD protein remains bound to the carboxylase and consequent processivity. Interestingly, the patient with the V255M and S300F mutations showed 33% and 64% clotting activities for FX and prothrombin, respectively,36 despite 100-fold reported differences in their EBD affinities.66 The patient’s phenotype suggests that processivity is not solely due to EBD affinity and likely involves additional mechanisms such as the transition after Glu carboxylation that allows the exchange of carboxylated and uncarboxylated VKD proteins (Figure 7B).
Acknowledgments
This work was supported by National Institutes of Health National Heart, Lung and Blood Institute and National Institute of Aging grants R01HL158007 and R01HL152678 (K.L.B. and K.W.R.) and NIH grant R01AG051601 and National Science Foundation grant 1908875 (K.W.R.).
Authorship
Contribution: M.A.R., K.W.H., L.A.W., and J.M.H. performed the experiments; M.A.R. and K.L.B. designed the research and analyzed the results; and M.A.R., K.L.B., and K.W.R. prepared the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Kathleen L. Berkner, Department of Cardiovascular and Metabolic Sciences, Cleveland Clinic Lerner College of Medicine, Lerner Research Institute, 9500 Euclid Ave, NB50, Cleveland, OH 44195; e-mail: berknek@ccf.org.
References
Author notes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

![Processive carboxylation of FIX is impaired in the V255M mutant. (A) A complex between the carboxylase and FIX was reacted in the presence of a challenge FIX (FIX−18/+41), and carboxylation of both FIX forms was monitored. (B-E) Independent gels were used to monitor FIX−18/+41 and fIX in the wild-type (WT) or V255M complexes because of differences in their size, and [14C]-standards were included on the gels to allow quantitation of carboxylation products. (F-G) FIX−18/+41 was carboxylated after FIX in the complex with WT carboxylase but occurred simultaneously with FIX in the complex with V255M carboxylase. FIX carboxylation was much lower in the complex with the V255M mutant than with WT carboxylase. It was also lower than observed in the reaction monitoring catalysis (Figure 2E), which may have been due to the lower KH2 concentrations used in the processivity assay. (H) S300F carboxylation of both FIX in the complex and FIX−18/+41 was poor.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/140/15/10.1182_blood.2021014275/8/m_blood_bld-2021-014275-gr3.jpeg?Expires=1763484263&Signature=rokaP~GkNAb37HqfUY5K~ZhUIoNk~FSMdzvgj4DRaN18Z5VOjFkIarQdgvaHsFO2Jp-WK-FWQuWZKtQGs9L6oR3gx5MWWGhqCDpT851QbBFmdMN2lwLxwi29N9VRMHsb2Ls-UC~rHPT-WzhjwYVc1R4RXog8lFPke~IEv~OeloYfh8b9M0r34zWgbdPuEsRxPUaWl216i3diRcqvntnzi9sWlVvKPFbjuL44i4BG8hi-m91h687i5gpD2dUE6MqTrzI852P7pOeZXNjgmeI3ArOxpvU4Mhn896Lmg2cNA8pdY3zqmutxA7Mx~HFNl5LRra9o9tosMk4PqTCkvYMZSw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Processive MGP carboxylation is impaired in the V255M and S300F mutants. (A) Complexes between MGPFLAG and variant carboxylases were reacted in the presence of a challenge peptide (MGP1-64) that contained both the EBD and the entire Gla domain. (B-D) MGPFLAG-carboxylase complexes were isolated on anti-FLAG resin, and the amount of complex was determined by an epoxidase assay, as before.20 Equivalent amounts of complex were then analyzed in the challenge assay. Carboxylation of MGPFLAG in the complexes and MGP1-64 was quantitated by monitoring [14C]-CO2 incorporation into each form, using gel electrophoresis and PhosphorImager analysis. (E-G) MGP1-64 carboxylation occurred after MGPFLAG in the complex with wild-type carboxylase. Both MGP forms were carboxylated at the same time by mutant carboxylases, which produced higher levels of MGP1-64 than MGPFLAG in the complex.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/140/15/10.1182_blood.2021014275/8/m_blood_bld-2021-014275-gr5.jpeg?Expires=1763484263&Signature=0zPpCt2nhCwc6aQ4CuRrsf1JYwhF9dpkr6AOGJAzmqGuZX3RegiTPh-ntluszW~TdntonoPN5ncp3yIbpqKmm3M33kuRfQmrDz6rbQEmyVd045W1JbESLvaKZ1NRlYGhGdqUEihnuA15vRwYxZ1WbH5akbJ3yjjH-6g9E3CcFgQ1IRgb4kCU-hz4N--1cJ~HxngwXf4goU7WUCqp3~ZJZPlOcW8KDTxXgJxyR5u58OfSk8uEnNkJNGDaCX14yBnJMjCzmbWzp7iI5~D7AV6~cZz1w4dbSmN50tbjbPJ~NngVj60RCuTP5CEB405drSC~VJQvH3jbe~3~oPG5ZWA74g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![The V255M mutant shows higher levels of modified MGP than wild-type (WT) carboxylase. (A) The overall reaction for MGP carboxylation comprises binding, catalysis, and release, which produces MGP containing 5 Glas (indicated by Ys). (B) MGP1-64 and FIX−18/+41 were incubated with the same amount of WT carboxylase, followed by SDS-PAGE and PhosphorImager analysis to quantitate [14C]-CO2 incorporation into the peptides. (C) The same assay was used to measure MGP1-64 carboxylation by WT and mutant carboxylases, using equivalent amounts of enzyme as determined by western blot analysis with an anti-carboxylase antibody. Higher carboxylase concentrations were used than in panel B, and the duplicate samples that were assayed gave almost identical results and consequent small error bars. The overall reaction for WT carboxylase took 60 minutes, as determined by comparing pmol of Gla in MGP1-64 with the amount of carboxylase. (D-F) The specific catalytic step was monitored using preformed MGPFLAG-carboxylase complexes generated in cells, which were adsorbed to anti-FLAG resin to remove free carboxylase. The amount of carboxylase was quantitated by an epoxidase assay,20 and equivalent amounts of complex were then incubated in reactions containing KH2 and [14C]-CO2, followed by gel electrophoresis and PhosphorImager analysis to monitor [14C]-CO2 incorporation into MGPFLAG.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/140/15/10.1182_blood.2021014275/8/m_blood_bld-2021-014275-gr6.jpeg?Expires=1763484263&Signature=wnyaVB9pbrn8nHObnyYvQSUiJvd2vLNexey-6tkQHvALSmJ5Cx1bSxtI~BeWyid2oi6vqIQNOUp9KZ45kq4ntiaGeiWB8CHeKHgcPsZgbeyi1azY4TfOKFXh14vK1yV21ITCp2zJ1VoGQdjfVE5cif9pnodtpT0kLU6Ls82umrf9nlvpNICzHu4xjwgn2J59k10EaIhrMl-eGQ0cJ4LsX2tmi6gmnpgbVEMKFuSKa5fTGMKort4-2mUEhsXAh6oivCZcLIw~~EMipjAE5Wti2COLjZQmp-DSHBszFzbcc0oskj9u0VQTnkkSUfoM0~5rJaU4WWmM7TehYCntVMLHaw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal