Key Points
CD34− MPP6 and CD34+ MPP5 (both LSK CD135−CD48−CD150−) are functionally located between HSCs and MPP2-4.
Transplanted MPP5 is important for emergency myelopoiesis and long-term lymphoid reconstitution.
Abstract
Hematopoietic stem cells (HSCs) and distinct multipotent progenitor (MPP) populations (MPP1-4) contained within the Lin−Sca-1+c-Kit+ (LSK) compartment have previously been identified using diverse surface-marker panels. Here, we phenotypically define and functionally characterize MPP5 (LSK CD34+CD135−CD48−CD150−). Upon transplantation, MPP5 supports initial emergency myelopoiesis followed by stable contribution to the lymphoid lineage. MPP5, capable of generating MPP1-4 but not HSCs, represents a dynamic and versatile component of the MPP network. To characterize all hematopoietic stem and progenitor cells, we performed RNA-sequencing (RNA-seq) analysis to identify specific transcriptomic landscapes of HSCs and MPP1-5. This was complemented by single-cell RNA-seq analysis of LSK cells to establish the differentiation trajectories from HSCs to MPP1-5. In agreement with functional reconstitution activity, MPP5 is located immediately downstream of HSCs but upstream of the more committed MPP2-4. This study provides a comprehensive analysis of the LSK compartment, focusing on the functional and molecular characteristics of the newly defined MPP5 subset.
Introduction
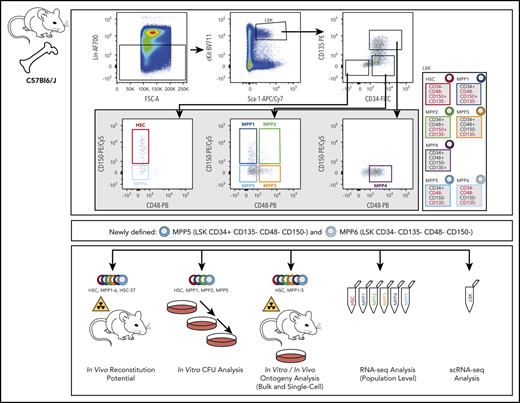
Multipotent hematopoietic stem cells (HSCs) are located at the top of the hematopoietic hierarchy and together with multipotent progenitors (MPPs)1-4 are contained within the Lin−Sca-1+c-Kit+ (LSK) compartment. Different markers have been used to identify and characterize mouse hematopoietic stem and progenitor cells (HSPCs), including CD34, CD135, CD48, and CD150.5-7 Although HSCs exhibit unique serial multilineage long-term engraftment potential, MPPs show a lower and variable level of self-renewal capacity and lineage-differentiation potential.8,9 We and others have functionally and molecularly characterized HSCs and MPP1-4 (supplemental Figure 1A, available on the Blood Web site).8-10 Furthermore, analysis of the CD48− and/or CD150− MPPs has been performed (eg, short-term HSC [HSCST]).5,10-13 However, a comprehensive characterization of the complete LSK compartment using CD34/CD135/CD48/CD150 as surface markers is lacking. Here, we assess the LSK compartment in mice (Figure 1A; supplemental Figure 1A) and determine the potential of CD34− MPP6 and CD34+ MPP5, previously underexplored progenitor populations.
Lineage potential of MPP5 cells and HSC/MPP ontogeny analysis. (A) Workflow of functional and molecular analysis of HSC and MPP cells. (B) Analysis of HSC/MPP transplantations. Workflow of transplantation approach. The relative percentage of donor contribution to the PB, myeloid, B-cell, and T-cell lineage is shown; n = 4-11. (C) Analysis of HSCST/MPP5/6 transplantations. Workflow of transplantation approach. The relative percentage of donor contribution to the PB, myeloid, B-cell, and T-cell lineage is shown; n = 5-11. (D) CFU assay of HSC, MPP1, MPP2, and MPP5 cells showing colony numbers for first, second, and third plating.; n = 6-9 (first, second); n = 2-5 (third). (E) In vitro ontogeny analysis. Frequency of single cells in the respective HSPC subset is shown 6 hours, 16 hours, 24 hours and 48 hours after plating as identified in flow cytometry analysis. Statistical analysis between the populations (respective color) or overall output (black) is shown compared with HSCs; n = 8 (48 hours: n = 3). (F) High-throughput simultaneous division and differentiation tracking per ancestor. Cells were stained with distinct carboxyfluorescein diacetate succinimidyl ester (CFSE)/CellTrace Violet (CTV) combinations and HSC/MPP populations were index sorted combining 4 cells of distinct cell trace colors per well. Twenty-four hours after culture, phenotypic identification was performed allowing analysis of differentiation trajectories from a founding ancestor. (G) In vivo ontogeny analysis. Endpoint analysis of recipient animals 1 or 2 weeks following transplantation in sublethally irradiated recipients. HSPC engraftment in the BM is shown as frequency of single cells. Statistical analysis between the populations (respective color) or overall engraftment (black) is shown compared with HSCs; n = 5-9. For all panels, mean plus standard deviation (SD) is shown. Two-way analysis of variance (ANOVA) (B,D-F). (B) Statistical analysis of MPP5 vs HSC is shown. *P < .05; **P < .01; ***P < .001. "n" indicates number of biological replicates. Two to 3 independent experiments (B-C), 2 to 4 independent experiments (D), 6/16/24 hours: 4 independent experiments; 48 hours: 1 independent experiment (E) 3 independent experiments (F-G). See also supplemental Figures 1 and 2. ns, not significant.
Lineage potential of MPP5 cells and HSC/MPP ontogeny analysis. (A) Workflow of functional and molecular analysis of HSC and MPP cells. (B) Analysis of HSC/MPP transplantations. Workflow of transplantation approach. The relative percentage of donor contribution to the PB, myeloid, B-cell, and T-cell lineage is shown; n = 4-11. (C) Analysis of HSCST/MPP5/6 transplantations. Workflow of transplantation approach. The relative percentage of donor contribution to the PB, myeloid, B-cell, and T-cell lineage is shown; n = 5-11. (D) CFU assay of HSC, MPP1, MPP2, and MPP5 cells showing colony numbers for first, second, and third plating.; n = 6-9 (first, second); n = 2-5 (third). (E) In vitro ontogeny analysis. Frequency of single cells in the respective HSPC subset is shown 6 hours, 16 hours, 24 hours and 48 hours after plating as identified in flow cytometry analysis. Statistical analysis between the populations (respective color) or overall output (black) is shown compared with HSCs; n = 8 (48 hours: n = 3). (F) High-throughput simultaneous division and differentiation tracking per ancestor. Cells were stained with distinct carboxyfluorescein diacetate succinimidyl ester (CFSE)/CellTrace Violet (CTV) combinations and HSC/MPP populations were index sorted combining 4 cells of distinct cell trace colors per well. Twenty-four hours after culture, phenotypic identification was performed allowing analysis of differentiation trajectories from a founding ancestor. (G) In vivo ontogeny analysis. Endpoint analysis of recipient animals 1 or 2 weeks following transplantation in sublethally irradiated recipients. HSPC engraftment in the BM is shown as frequency of single cells. Statistical analysis between the populations (respective color) or overall engraftment (black) is shown compared with HSCs; n = 5-9. For all panels, mean plus standard deviation (SD) is shown. Two-way analysis of variance (ANOVA) (B,D-F). (B) Statistical analysis of MPP5 vs HSC is shown. *P < .05; **P < .01; ***P < .001. "n" indicates number of biological replicates. Two to 3 independent experiments (B-C), 2 to 4 independent experiments (D), 6/16/24 hours: 4 independent experiments; 48 hours: 1 independent experiment (E) 3 independent experiments (F-G). See also supplemental Figures 1 and 2. ns, not significant.
Study design
For detailed methods, see supplemental Methods.
Mice
C57BL/6J (CD45.2, .1 or .1/.2) mice were used. All animal procedures were performed according to protocols approved by the German authorities, Regierungspräsidium Karlsruhe (Nr. A-23/17, Z110/02, DKFZ 299, G-140/13. G-183/17).
Cell suspensions and flow cytometry
BM cells were isolated, HSPCs were purified by FACS (Aria I, II, III or FACS Aria Fusion; Becton Dickinson), and subsequently subjected to in vivo or in vitro assays or RNA-Seq analysis. For scRNAseq analysis, LSK cells were sorted. Antibody conjugates were purchased from eBioscience or BioLegend.
Reconstitution experiments
Two thousand cells per population were sorted and transplanted into fully irradiated (2 × 5 Gy) B6 mice (CD45.1) together with 2 × 105 total spleen cells (CD45.1/.2). The CD45.2 contribution was monitored in PB. For endpoint analysis, BM and spleen stainings were performed. For secondary transplantations, whole BM was isolated 16 weeks after transplantation and 3 × 106 cells were retransplanted.
Colony-forming unit (CFU) assays
One thousand FACS-sorted cells were cultured in MethoCult M3434 (StemCell Technologies) in technical replicates (500 cells per plate). After 7 days, colony formation was quantified and 30 000 cells were replated and quantified 5 days later for secondary and tertiary platings.
In vitro ontogeny assay
Two thousand cells per analysis time point (6, 16, 24, and 48 hours) were sorted and cultured in complete stem cell medium and subsequently stained for flow cytometry-based analysis.
RNA-seq and scRNAseq sequencing
RNA sequencing (RNA-seq) data of sorted MPP5 cells was generated and single-cell RNA-seq (scRNAseq) analysis of LSK cells was performed. See additional information provided in the supplemental Methods. RNA-seq (MPP5) and scRNAseq (LSK) data are available under ArrayExpress (E-MTAB-9135 and E-MTAB-9208).
Results and discussion
We partitioned the LSK compartment (supplemental Figure 1A), enabling the definition of MPP5 (LSK CD34+ CD135−CD48−CD150−) and MPP6 (LSK CD34−CD135−CD48−CD150−). Comparison of MPP5/MPP6 and MPP1-4, as previously defined,9 to MPP populations defined by Pietras et al10 (lacking CD34) and Oguro et al11 (lacking CD34/CD135), revealed a high overlap between MPP2-4 compartments, when using CD135 but not CD34 (supplemental Figure 1B, top panel) and a mixture of MPPs when both markers were not included (supplemental Figure 1B, bottom panel). Importantly, CD34 added an additional layer of information, as the previously defined long-term HSC (HSCLT)/HSCST (LSK CD135−CD150+/−CD48−)10 compartments could be further subdivided into a CD34− and CD34+ subpopulation (CD34− HSC/CD34+ MPP1, CD34− MPP6/CD34+ MPP5). As serial multilineage engraftment is characteristic for CD34− cells, this subdivision is important to isolate populations with the highest self-renewal capacity.6,9
The self-renewal and differentiation capacity of all HSPC populations was assessed by reconstitution analyses in vivo (Figure 1B-C). HSCs displayed the expected long-term multilineage engraftment (Figure 1B) by peripheral blood (PB) analysis. In contrast, MPP5 exhibited very unique engraftment patterns with very early but transient myeloid cell production and subsequent stable long-term contribution to lymphoid lineages consistent with previously defined CD48− and/or CD150− subsets (but excluding CD34/CD135 and/or CD48)5,10-12,14 but different in terms of transplantation dynamics. Regarding overall contribution to PB and myeloid lineages, MPP5 exhibited the highest cellular output shortly after transplantation compared with all other populations (9/10d), suggesting a role as an emergency reservoir, at least in the setting of transplantation. The contribution of transplanted MPP1-4 cells to PB was comparable to previously published studies.9,10 Comparison with described CD48−CD150− cell subsets (HSCST)10 revealed differences in engraftment capacity and lineage contribution (Figure 1C). MPP5 and MPP6 show distinct PB contribution, with MPP5 (CD34+) showing fast short-term myeloid and long-term lymphoid engraftment, whereas MPP6 (CD34−) displays slower dynamics but shows long-term multilineage contribution similar to HSCs 4 to 12 weeks posttransplantation. In agreement, previously reported HSCST,10 which includes MPP5 as well as MPP6 cells (supplemental Figure 1B, top panel), exhibits a mixed MPP5/MPP6 engraftment pattern with early engraftment mimicking MPP5 and long-term engraftment mimicking MPP6 patterns. These findings underline the importance of using CD34 as a marker to distinguish progenitor populations with long-term multilineage potential.
To further analyze MPP5, we next characterized the reconstitution capacity of MPP5 in the bone marrow (BM). HSC transplantation led to full replenishment of all HSPCs, whereas MPP5 revealed stable engraftment of itself and MPP3/MPP4 (supplemental Figure 1C). In the BM and spleen of MPP5 recipients, high contribution to lymphoid lineages was detected, but few myeloid cells were detected (supplemental Figure 1D-E). In secondary recipients (supplemental Figure 1F), only very minor MPP5-derived contributions were found, suggesting a limited self-renewal activity confirming that serial multilineage engraftment is restricted to CD34− HSCs.6,9 The lymphoid cells observed in secondary MPP5 transplants could potentially also be generated by more committed progenitors derived from MPP5 cells in primary recipients. MPP5 seems to act as a reservoir to quickly replenish the myeloid compartment in emergency situations, whereas they are able to produce B and T cells long-term in primary recipients. CFU analysis mirrored the results obtained from transplantations, with HSCs showing the highest serial colony-forming potential and MPP5 showing higher potential only in primary platings (Figure 1D), supporting the rationale that MPP5 is primed for emergency myeloid output associated with restricted self-renewal activity.
To better understand the ontogeny within the LSK compartment, we performed short-term in vitro differentiation analyses (Figure 1E; supplemental Figure 1G). As expected, HSCs were only detected in HSC cultures and gave rise to MPP1, confirming their position at the top of the hierarchy. MPP1 was maintained or showed additional MPP2/MPP5 marker expression patterns, whereas MPP2 mainly preserved their fate and only a few MPP3/MPP4 were detected. MPP3 and MPP4 exhibited a mixed MPP3/MPP4 potential, giving rise to the respective population. In contrast to MPP1-4, MPP5 gave rise to all other MPPs, identifying them as a hub within the MPP network. These findings are in line with previous reports10,15 as well as our in vivo results suggesting that at least in vitro, MPP5 can serve as master-type MPPs (supplemental Figure 1H).
To refine the ontogeny at the single-cell level, we traced the progeny cell types from single-cell ancestors in vitro16 and obtained similar results compared with bulk analysis, confirming the broad differentiation capacity of MPP5 (Figure 1F; supplemental Figure 2A). In addition, the fraction of cells remaining in the LSK gate was higher in HSC, MPP1, and MPP5 than in MPP2-4, both in bulk and single-cell analysis, in line with their position at the top of the hierarchy.
To investigate MPP hierarchies in vivo, we transplanted purified HSC and MPP cells in sublethally irradiated recipient animals and performed BM analysis after 1 or 2 weeks (Figure 1G; supplemental Figure 2B). Interestingly, a high degree of flexibility within the MPP network was observed in this condition of transplantation stress. In line with our proposed in vitro differentiation model (supplemental Figure 1H), MPP populations could give rise to one another. However, HSCs could only be observed in HSC-recipient animals, again underlining their position at the top of the hematopoietic hierarchy. Although there is a high amount of differentiation flexibility in vivo within the MPP compartment, transplantation dynamics are different between MPPs. As expected, MPP5 cells showed the highest and most balanced MPP contribution to total BM after 1 week, whereas their expansion decreased significantly after 2 weeks. Characteristic engraftment levels, dynamic expansion, and varying sizes in the different MPP pools were observed for all analyzed HSPC compartments. These differences in dynamics and the potential to give rise to different numbers of progeny cells of different MPPs in vivo, which in turn have a defined lifespan and self-renewal capacity, presumably lead to the distinct engraftment patterns observed upon long-term transplantation of the different MPP populations (Figure 1B). The provided in vivo data are in agreement with our in vitro analysis. However, the approach relies on surface marker profiles and in vivo potential was analyzed in the setting of stress, for example, irradiation. Additional molecular analysis of the transplanted populations and progeny cells would be necessary to exclude potential uncertainties regarding the molecular identity of the analyzed cells. The limited number of recoverable cells using this experimental approach currently limits molecular analysis. Thus, for the moment, the suggested in vivo differentiation potential has to be regarded with caution and future studies need to further substantiate our proposed model.
To better understand the molecular differences within HSPCs focusing on MPP5, we made use of our previously generated transcriptome data set adding novel equivalent data on MPP5 and reanalyzed the entire data set (Figure 2A).17 Unsupervised clustering grouped HSC/MPP1/MPP5 and MPP2/MPP3/MPP4 together (supplemental Figure 3A). This is in agreement with in vivo data, as HSCs, MPP1, and MPP5 robustly engraft in recipients (Figure 1).9,10 Principal component analysis further underscored these findings (Figure 2B). Across all populations, we identified 3405 differentially expressed genes (DEGs) and confirmed RNA-seq expression patterns by quantitative reverse transcription–polymerase chain reaction (supplemental Figure 3B; supplemental Table 1). Pairwise comparison revealed ∼2000 DEGs between HSCs and MPP2/3/4, and only 1098 DEGs between MPP1 and HSCs (Figure 2C). Molecular profiling of MPP5 revealed 1034 DEGs compared with HSCs. Fewer than 1000 DEGs were identified between MPP5 and MPP3/MPP4, and only 227 DEGs comparing MPP5 to MPP1, providing additional evidence that MPP5 may function as a hub within the MPP network. We assigned each of the DEGs to 1 of the HSPC populations to generate gene signatures of HSCs and MPP1-5 as previously described9 (Figure 2D; supplemental Table 2). Concurring with their in vivo potential, analysis of hallmark gene sets between MPP5 and HSCs revealed enrichment in cell cycle–related pathways, metabolic processes like oxidative phosphorylation, and inflammatory responses (Figure 2E). Interestingly, processes enriched in MPP1 compared with HSCs also included cell cycle–associated and metabolic processes, but lacked inflammatory pathways. Consequently, inflammatory processes were enriched in MPP5 compared with MPP1, whereas MPP1 exhibited even higher enrichment in cell-cycle pathways, highlighting the different molecular characteristics of these 2 closely related cell populations (Figure 2E). In contrast to other MPPs,17 the close relationship of MPP5 and HSCs was also observed at the level of alternative polyadenylation as only 3 differentially used sites and a modest shortening of 3′ untranslated regions could be identified (supplemental Figure 3C-D).
Molecular analysis of HSPCs and LSK cells on the population and single-cell level. (A) Workflow generation population RNA-seq data. (B) Principal component analysis. (C) Relative distances are shown based on numbers of DEGs in pairwise comparisons. (D) Gene-expression clusters. DEGs were grouped into 6 clusters based on higher expression (enriched) in the respective cell population compared with mean expression level across all cell populations. Number of genes in the respective gene cluster is shown. (E) Hallmark-term analysis of gene sets enriched in MPP5 compared with HSCs, HSCs compared with MPP1 and MPP1 compared with MPP5. Normalized enrichment score (NES) for significant pathways (adjusted P [padj] < .05) is shown. (F) Workflow generation scRNAseq data. (G) Uniform manifold approximation and projection (UMAP) projection. Clusters are color-coded based on enrichment scores, and clusters were associated to the respective gene signatures identified in the population RNA-seq analysis (based on panel H). (H) Expression of HSPC gene signatures in the respective clusters. Red circles indicate maximal enrichment ± 0.02 considering the signature scoring. Analysis of maximal enrichment was used to assign colors and MPP identity to the respective clusters. (I) UMAP projection. Coloring is derived from the cell-cycle status. (J) UMAP projection. Trajectory analysis is depicted. (K) Pseudotime analysis. Zoom in in gray box shows analysis for selected clusters 0, 1, 2, 4, and 6. (L) Workflow in vitro barcoding and single-cell (sc) profiling,27 Clone label transference within HSPC clusters between 2 and 4 days of in vitro culture was analyzed based on the respective gene signatures identified in the population RNA-seq data. (M) Workflow in vitro barcoding and in vitro/in vivo single-cell profiling.27 Fate probability matrix is shown. HSPC identity of clones (2 days) was identified based on population RNA-seq gene signatures and lineages were assigned based on marker expression (2-3 weeks after transplantation). See also supplemental Figures 3 and 4. DC, dendritic cell; FACS, fluorescence-activated cell sorting; Mk/Ery, megakaryocyte/erythrocyte; Mono, monocyte; Neu, neutrophil; QC, quality control.
Molecular analysis of HSPCs and LSK cells on the population and single-cell level. (A) Workflow generation population RNA-seq data. (B) Principal component analysis. (C) Relative distances are shown based on numbers of DEGs in pairwise comparisons. (D) Gene-expression clusters. DEGs were grouped into 6 clusters based on higher expression (enriched) in the respective cell population compared with mean expression level across all cell populations. Number of genes in the respective gene cluster is shown. (E) Hallmark-term analysis of gene sets enriched in MPP5 compared with HSCs, HSCs compared with MPP1 and MPP1 compared with MPP5. Normalized enrichment score (NES) for significant pathways (adjusted P [padj] < .05) is shown. (F) Workflow generation scRNAseq data. (G) Uniform manifold approximation and projection (UMAP) projection. Clusters are color-coded based on enrichment scores, and clusters were associated to the respective gene signatures identified in the population RNA-seq analysis (based on panel H). (H) Expression of HSPC gene signatures in the respective clusters. Red circles indicate maximal enrichment ± 0.02 considering the signature scoring. Analysis of maximal enrichment was used to assign colors and MPP identity to the respective clusters. (I) UMAP projection. Coloring is derived from the cell-cycle status. (J) UMAP projection. Trajectory analysis is depicted. (K) Pseudotime analysis. Zoom in in gray box shows analysis for selected clusters 0, 1, 2, 4, and 6. (L) Workflow in vitro barcoding and single-cell (sc) profiling,27 Clone label transference within HSPC clusters between 2 and 4 days of in vitro culture was analyzed based on the respective gene signatures identified in the population RNA-seq data. (M) Workflow in vitro barcoding and in vitro/in vivo single-cell profiling.27 Fate probability matrix is shown. HSPC identity of clones (2 days) was identified based on population RNA-seq gene signatures and lineages were assigned based on marker expression (2-3 weeks after transplantation). See also supplemental Figures 3 and 4. DC, dendritic cell; FACS, fluorescence-activated cell sorting; Mk/Ery, megakaryocyte/erythrocyte; Mono, monocyte; Neu, neutrophil; QC, quality control.
To address heterogeneity, we performed scRNAseq of 5520 LSK cells (Figure 2F) forming 7 clusters (Figure 2G; supplemental Table 3). Using our gene signatures (Figure 2D), we associated the scRNAseq clusters to the different HSPCs (Figure 2G-H; supplemental Figure 3E) and confirmed this approach using a previously published scRNAseq LSK data set18 (supplemental Figure 3F). Analysis of reported HSC signatures (MolO; NoMO)19 and expression patterns of HSPC markers and regulators9,10,20,21 further established the identity of the clusters (supplemental Figures 3G-H and 4A). In addition, we identified markers and potential regulators of MPP5 clusters (# 0, 4, 6) (supplemental Figure 4B). This includes histone modifiers such as the peptidyl arginine deiminase 4 (Padi4) and the demethylase Kdm7a, involved in chromatin regulation,22,23 as well as Nfkbiz, involved in inflammatory processes.24 Cell-cycle scores and regulators (Cdk6, Mki67) exhibited the expected distribution,8,25,26 with HSC/MPP1/MPP5 localized mainly in G0/G1 clusters and MPP2/3/4 enriched for G2/M and S signatures as confirmed by flow cytometry analysis (Figure 2I; supplemental Figure 4C-E). Using HSCs (cluster 2) as a defined starting point, trajectory analysis was performed revealing the MPP1/MPP5 clusters to be localized immediately downstream of HSCs (Figure 2J). MPP5 were centrally located, with trajectories toward all other MPPs. MPP2/3/4 were closely connected, but were located more downstream without direct trajectory to HSCs. These data provide a molecular explanation for the ontogeny patterns and are further supported by pseudotime analysis (Figure 2K). The latter also confirmed HSCs (cluster 2) as the molecular starting point of the differentiation cascade, giving first rise to MPP1/MPP5 and subsequently to the MPP2/3/4 clusters.
To further analyze HSC-MPP trajectories, we reanalyzed published data27 in which LSK cells were sorted and barcoded using the LARRY system and single-cell profiling was performed 2 and 4 days after in vitro culture. Using our gene signatures to assign cell identities, we quantified label transfer between the different HSPC populations in vitro (Figure 2L; supplemental Figure 4F). The trajectories identified by this analysis are very much in line with our results based on in vitro surface marker profiling (Figure 1E-F). HSCs localize at the top of the hierarchy and MPP5 cells exhibit a high degree of label transfer to MPP2/3/4 clusters, whereas MPP2/3/4 mostly retain the label in their compartments (Figure 2L; supplemental Figure 4F). In addition, we reanalyzed MPP in vivo fate data 2 to 3 weeks following transplantation from the same study (Figure 2M).27 Here, we successfully projected our HSPC gene signatures (Figure 2D) on single-cell clusters at the starting point of the analysis (day 2) and quantified the average fate distribution observed in vivo for each HSPC population (Figure 2M; supplemental Figure 4G). These data are not directly comparable to analysis of fate by surface marker profiling (Figure 1B) as PB was not analyzed 2 to 3 weeks after transplantation. However, we could confirm the balanced contribution of MPP5 cells to all of the different lineages analyzed at this early timepoint. Other MPP subsets showed less balanced lineage contribution (Figure 2M), in agreement with our PB analysis (Figure 1B). By using gene signatures and different single-cell sequencing data sets derived from freshly isolated LSK, and in vitro cultured and in vivo transplanted barcoded HSPCs, we were able to provide a first definition of trajectories within the HSPC compartment and identify MPP5 as a myeloid source for emergency situations such as irradiation.
We report a close relationship between MPP1 and MPP5, placing them downstream of HSCs but upstream of less potent MPP2/3/4. MPP5 forms a dynamic MPP cloud and MPP6 represents a subset with higher multilineage potential compared with MPP5. Our suggested partitioning of HSPCs, which is linked to the biological and functional potential of cells, should be understood as a complement to models derived from high-resolution, but descriptive, single-cell omics data. Although the latter can provide explanations for the continuous changes occurring during differentiation at the single-cell level, surface marker–based identification of cells remains currently the only way to isolate and functionally characterize single cells and populations. In this study, together with the already reported characterization of MPP1-4, we functionally characterize MPP5 and MPP6 and provide the transcriptomic landscapes of MPP1-5, further refining the definition of progenitor populations downstream of HSCs within the classical LSK gate. Future molecular characterization of MPP6 will help to integrate it into the HSC/MPP1-5 differentiation trajectories and potentially reveal novel and distinct differentiation pathways such as distinct routes via the MPP1/2 and MPP5/6 families.
RNA-seq data and scRNAseq data reported in this article are available in ArrayExpress (E-MTAB-9135 and E-MTAB-9208). Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Andreas Trumpp (a.trumpp@dkfz.de). Certain materials are shared with research organizations for research and educational purposes only under a material transfer agreement to be discussed in good faith with the recipient.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank M. Sohn, A. Przybylla, and all technicians of the A.T. laboratory for technical assistance; S. Schmitt, M. Eich, K. Hexel, T. Rubner, and F. Blum from the German Cancer Research Center (DKFZ) Flow Cytometry Core Facility for their assistance; and K. Reifenberg, P. Prückl, M. Durst, M. Schorpp-Kistner, A. Rathgeb, and all members of the DKFZ Laboratory Animal Core Facility for excellent animal welfare and husbandry. The authors also thank the DKFZ Genomics and Proteomics Core Facility for their assistance, as well as M. Derecka, D. Grün, and E. Trompouki for careful reading of the manuscript.
This work was supported by the Max Planck Society, a 2017 European Research Council starting grant (ERC-Stg-2017) (VitASTEM; 759206), the German Research Foundation (DFG) under Germany’s Excellence Strategy (CIBSS-EXC-2189–Project ID 390939984), and the European Union’s Horizon 2020 Research and Innovation Programme under the Marie Skłodowska-Curie Grant (agreement no. 813091) (all to N.C.-W.). This work was also supported by DFG grants FOR2033, FOR2674, and SFB873, German Cancer Consortium (DKTK) joint funding “RiskY-AML,” and the SyTASC Consortium funded by the Deutsche Krebshilfe, and the Dietmar Hopp Foundation (all to A.T.). S.A. acknowledges funding from the European Hematology Association (EHA; Advanced Nonclinical Research Fellowship). A.R.-F. is a scholar of the American Society of Hematology and a special fellow of the Leukemia & Lymphoma Society.
Authorship
Contribution: P.S., N.C.-W., and A.T. designed the experiments and wrote the manuscript; P.S., L.L., L.H., A.N., K.S., S.R., P.Z., K.J., and D.K. performed experiments; P.S., M.C.R.-M., A.N., S.A., A.R.-F., L.H., and L.L. analyzed the data; L.P., F.D.C., M.C.R.-M., A.N., and L.L. provided comments on the manuscript; and N.C.-W. and A.T. supervised all experiments.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Nina Cabezas-Wallscheid, Max Planck Institute of Immunobiology and Epigenetics, Stübeweg 51, D-79108 Freiburg, Germany; e-mail: cabezas@ie-freiburg.mpg.de; and Andreas Trumpp, DKFZ, Im Neuenheimer Feld 280, D-69120 Heidelberg, Germany; e-mail: a.trumpp@dkfz.de.
REFERENCES
Author notes
P.S., M.C.R.-M., A.N., and L.L. contributed equally to this study.
A.T. and N.C.-W. are joint senior authors.


![Molecular analysis of HSPCs and LSK cells on the population and single-cell level. (A) Workflow generation population RNA-seq data. (B) Principal component analysis. (C) Relative distances are shown based on numbers of DEGs in pairwise comparisons. (D) Gene-expression clusters. DEGs were grouped into 6 clusters based on higher expression (enriched) in the respective cell population compared with mean expression level across all cell populations. Number of genes in the respective gene cluster is shown. (E) Hallmark-term analysis of gene sets enriched in MPP5 compared with HSCs, HSCs compared with MPP1 and MPP1 compared with MPP5. Normalized enrichment score (NES) for significant pathways (adjusted P [padj] < .05) is shown. (F) Workflow generation scRNAseq data. (G) Uniform manifold approximation and projection (UMAP) projection. Clusters are color-coded based on enrichment scores, and clusters were associated to the respective gene signatures identified in the population RNA-seq analysis (based on panel H). (H) Expression of HSPC gene signatures in the respective clusters. Red circles indicate maximal enrichment ± 0.02 considering the signature scoring. Analysis of maximal enrichment was used to assign colors and MPP identity to the respective clusters. (I) UMAP projection. Coloring is derived from the cell-cycle status. (J) UMAP projection. Trajectory analysis is depicted. (K) Pseudotime analysis. Zoom in in gray box shows analysis for selected clusters 0, 1, 2, 4, and 6. (L) Workflow in vitro barcoding and single-cell (sc) profiling,27 Clone label transference within HSPC clusters between 2 and 4 days of in vitro culture was analyzed based on the respective gene signatures identified in the population RNA-seq data. (M) Workflow in vitro barcoding and in vitro/in vivo single-cell profiling.27 Fate probability matrix is shown. HSPC identity of clones (2 days) was identified based on population RNA-seq gene signatures and lineages were assigned based on marker expression (2-3 weeks after transplantation). See also supplemental Figures 3 and 4. DC, dendritic cell; FACS, fluorescence-activated cell sorting; Mk/Ery, megakaryocyte/erythrocyte; Mono, monocyte; Neu, neutrophil; QC, quality control.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/137/23/10.1182_blood.2020007876/5/m_bloodbld2020007876f2.png?Expires=1767706873&Signature=1lZxSZmsUML8dIBuAVwVKev~0Iy~8hO1tIR-PcYC0-hU848n5xNlRBn8QARwEswhW1Aj--P~jW3xaOoj7~1EOwGde9WgA0voRCopgw7dHcpKak07b~ObwZBQqQGHtdy9qoUyCNJAvRSi5OhqpVC9cL2LmC~dewH6lRtUpFFmzgDVBVe4LZUo6qkIiOKTqfxdqwznzEPcWyTouEv8mn9FeFhJ-judH97RIyX96ruZWPyJzx-sqeUh1EbW5R0hck6gY5Yuj61bsMum-w23WorN68WFJnxHG6Z-GJXHks6GF3bwzsX8Tp5vfW97ZgNlgBaPuAiG5UATbEyyRmOm07z6rg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal