In this issue of Blood, Zinter et al report results of metatranscriptomic sequencing of bronchoalveolar lavage fluid (BALF) from asymptomatic pediatric hematopoietic cell transplantation (HCT) candidates.
Using metatranscriptomics from BALF to predict posttransplantation lung injury in pediatric HCT candidates: innovations and challenges.
Using metatranscriptomics from BALF to predict posttransplantation lung injury in pediatric HCT candidates: innovations and challenges.
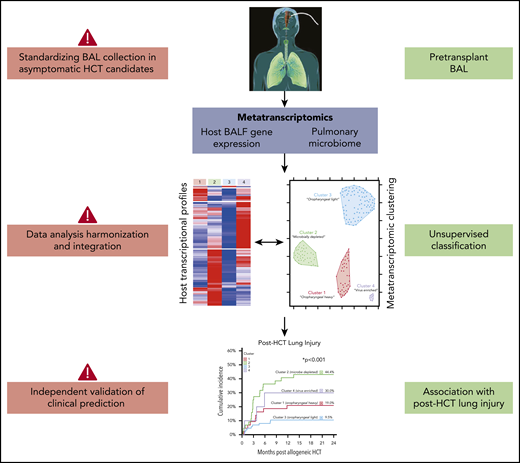
Zinter et al correlate microbe-host gene expression clusters with posttransplant lung injury, a clinically significant outcome associated with high morbidity and mortality.1 The investigators used a large set of highly unique clinical samples and applied parallel and integrative analyses of host gene expression and microbiome data to identify specific metatranscriptomic clusters associated with posttransplant outcomes. The main finding from this innovative approach was that pediatric HCT candidates with viral enrichment, microbial depletion, immune activation, and response to cellular injury are more likely to develop posttransplant lung injury. The authors suggest that identification of high-risk patients through pretransplant BALF metatranscriptomic analysis may open a window of opportunity for targeted therapy that can influence posttransplant outcomes.
These findings are consistent with several lines of evidence that indicate the detrimental impact of pretransplant respiratory viral infections on long-term pulmonary outcomes in both pediatric and adult patients. Interestingly, in contrast to other studies,2-4 detection of rhinovirus, by far the most common respiratory virus detected in HCT candidates and recipients, was not associated with poor posttransplant clinical outcomes, perhaps because only asymptomatic individuals were included in this study. Regardless, given the high rate of respiratory viral detection in pediatric HCT candidates, additional evaluation of the relationship between detection of specific viruses in the context of microbiome composition and host inflammation may assist in refining protocols to delay transplantation or provide targeted therapy, even in asymptomatic individuals.
By combining microbial metagenomic sequencing with host gene expression data, Zinter et al provide a more comprehensive look into the interplay among the microbiome, respiratory pathogens, and immunity and investigate the impact of these interactions on clinical outcomes. Most studies in the HCT population have evaluated the gut microbiome for this purpose, with several studies suggesting that alterations in the intestinal microbiota are associated with poor posttransplant outcomes, including the development of severe respiratory viral disease.5-7 Evaluation of local host-microbe interactions is therefore highly relevant and represents a novel approach for understanding the potential mechanisms driving the pathogenesis of posttransplant lung injury.
For meaningful use in the clinical setting, several challenges with interpretation and application of BALF metatranscriptomics must be addressed (see figure). First, rigorous validation studies with both pediatric and adult cohorts are needed to confirm the association between the identified metatranscriptome clusters and lung injury. If validation studies consistently identify similar biologically plausible signatures, the next step in the field would be, as the Zinter et al suggest, to identify specific pathways for interventions that positively affect patient outcomes. Potential approaches include pathogen-directed therapy (ie, against respiratory viral pathogens), agents to improve local microbial diversity (ie, with pre- or probiotics), and host immune modifiers. In addition, such data could simply be used to inform rational decisions about delaying transplantation. Ideally, these approaches would be evaluated in well-designed clinical trials with clearly defined time points for intervention and appropriate clinical end points. Second, pretransplant BALF is not routinely obtained in asymptomatic HCT candidates. Most often, bronchoalveolar lavages (BALs) are performed to identify an infectious agent causing either symptoms or radiographic changes, especially in pediatric patients, in whom BALs are performed at a lower frequency than in the adult population. Changing this longstanding clinical practice can have its obstacles. Importantly, there is increasing evidence that bulk and single-cell RNA sequencing of peripheral blood can be leveraged to assess pulmonary disease severity in patients with respiratory viral infections, including influenza, respiratory syncytial virus, and SARS-CoV-2.8-10 If appropriate validation studies are performed that include immunocompromised populations, transcriptomic signatures from peripheral blood may represent a more practical tool for predicting lung injury. Similarly, obtaining specimens from more accessible locations, such as the upper respiratory or gastrointestinal tract, may be a more pragmatic approach to evaluating potential associations between the microbiome and outcomes.
Additional challenges remain in the use of complex high-dimensional data, such as host transcriptomic and microbiome sequencing data. Data acquisition, including timing and method of sample collection, and analysis pipelines should be harmonized to generate accurate, reproducible data that have biologic and potential clinical significance. Data harmonization is another reason that independent validation studies are paramount for extending the findings of Zinter et al. The validation cohort presented in this study takes the first step by confirming the metatranscriptomic clusters established in the primary cohort; however, outcome validation is also needed. Furthermore, currently, the cost and time needed to generate and analyze large-scale sequencing data are substantial, and the analysis requires significant expertise. Thus, the predictive value of metatranscriptomic data should be compared with known risk factors for the outcome of interest in order to critically evaluate the added benefit of performing complex and costly molecular fingerprinting.
Overall, Zinter et al have demonstrated the prognostic utility of assessing pulmonary host-microbial signatures from pretransplant BALF on posttransplant clinical outcomes in a high-risk pediatric population. Although there are several limitations in the generalizability of their approach for clinical applications, this study represents an exciting step in moving the field toward multidimensional pretransplant risk assessment, which may lead to targeted, preventative interventions that reduce serious pulmonary morbidities in patients who undergo HCT.
Conflict-of-interest disclosure: A.W. has served as an advisory board member for Kyorin Pharmaceutical, received institutional support from Ansun Biopharma and Allovir Inc for participation in sponsored research, and has received research support from VB Tech and Amazon Inc, all unrelated to this work. S.A.G. declares no competing financial interests.