In this issue of Blood, Fidanza et al elegantly describe the single-cell transcriptome of hematopoietic stem and progenitor cells (HSPCs) derived from human pluripotent stem cells (hPSCs) and compared it with human fetal liver progenitors using an artificial neural network.1
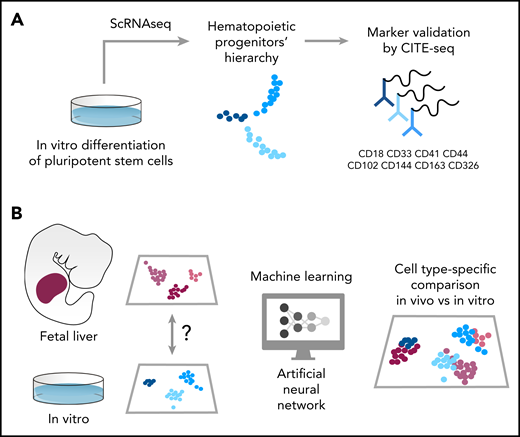
Schematic of the pipeline used in Fidanza et al to characterize hematopoietic cells differentiated from hPSCs. (A) The authors sequenced >40 000 cells in 2 experiments. The first scRNAseq experiment identified the transcriptome of uncommitted and lineage primed progenitors. The membrane markers associated with these cells were used to functionally validate their in silico–predicted lineage output. In the second experiment, 8 membrane markers were tagged using oligo-barcoded antibodies, and the cells were then sequenced using a CITE-seq approach. This allowed verification of the expression pattern of specific markers and associated them with a single-cell transcriptome. (B) The authors compared the single-cell transcriptome of hematopoietic cells collected from the human fetal liver with that of cells differentiated in vitro using a machine learning approach. First, they trained an artificial neural network using a vast single-cell fetal liver dataset and then used this trained network to transfer cell-type labels to the in vitro–derived cells. Finally, corresponding cell types were compared, and differentially expressed genes were listed as targets to improve the production of specific hematopoietic population in vitro from human PSCs.
Schematic of the pipeline used in Fidanza et al to characterize hematopoietic cells differentiated from hPSCs. (A) The authors sequenced >40 000 cells in 2 experiments. The first scRNAseq experiment identified the transcriptome of uncommitted and lineage primed progenitors. The membrane markers associated with these cells were used to functionally validate their in silico–predicted lineage output. In the second experiment, 8 membrane markers were tagged using oligo-barcoded antibodies, and the cells were then sequenced using a CITE-seq approach. This allowed verification of the expression pattern of specific markers and associated them with a single-cell transcriptome. (B) The authors compared the single-cell transcriptome of hematopoietic cells collected from the human fetal liver with that of cells differentiated in vitro using a machine learning approach. First, they trained an artificial neural network using a vast single-cell fetal liver dataset and then used this trained network to transfer cell-type labels to the in vitro–derived cells. Finally, corresponding cell types were compared, and differentially expressed genes were listed as targets to improve the production of specific hematopoietic population in vitro from human PSCs.
The ultimate challenge in the field of developmental hematopoiesis is understanding the complex differentiation landscape of HSPCs in order to reproduce in vitro, and eventually in vivo, the proper environmental cues to support long-term, multilineage hematopoietic stem cells (HSCs). Blood progenitors appear in the embryo in distinct hematopoietic waves, which differ from each other by their lineage output.2 Identification of specific cellular markers to isolate these progenitors is essential to explore the hierarchical relationships during developmental hematopoiesis. For this reason, integrative bioinformatics approaches are being used to improve our understanding of the cellular and molecular factors associated with the emergence of HSCs in vivo and in vitro.3
Fidanza et al reported an elegant, well-conducted study in which human induced pluripotent stem cells–derived HSPCs were sequenced and compared at the single-cell level with in vivo counterparts. More than 40 000 cells were sequenced in the experiments. In the first experiment, the authors selected the CD235a−CD43+ suspension cell population in order to exclude progenitors originating from the primitive wave. This heterogeneous mix of progenitors nicely clustered into clearly separable populations of uncommitted, immature progenitors and cells committed to megakaryocyte, erythroid, and granulocyte lineage, with the identity of each population marked by a unique repertoire of expressed genes. With a combination of semisolid clonogenic assays and a chimeric coculture system, the authors functionally validated a population of naïve progenitors characterized by the expression of CD44, a membrane protein involved in the endothelium-to-hematopoietic transition (EHT). This is especially important because CD44 low expression marks the onset of EHT, and it increases toward the emergence of HSCs.4 Among the membrane markers that characterize the hierarchy of in vitro hPSC-derived HSPCs, the authors found that CD326, CD41, and CD18 marked erythroid, megakaryocyte, and granulocyte lineage, respectively. These markers, among others, were further confirmed by a CITE-seq analysis in which cells were labeled with oligonucleotide-tagged antibodies specific for the above-mentioned markers (see figure). Interestingly, both suspension (CD235a−CD43+) and adherent fraction (CD235a−CD43−) of differentiating cells were included in the CITE-seq analysis. This is important because early blood progenitors were found among the adherent hemogenic endothelium, a cell population with hematopoietic potential that expresses endothelial markers such as CD31 (PeCAM1) and CD144 (VE-Cad).5,6 This approach tested the specificity of a minimal marker repertoire in capturing the early stages of lineage commitment (see figure).
A major strength of this work is the way the single-cell data have been compared with another landmark work on developmental hematopoiesis. This is the first time that machine learning has been used to compare in vitro– and in vivo–generated cell types. An artificial neural network7 was first trained using a published fetal liver dataset8 and then used to “recognize” cell types in the in vitro dataset (see figure). The authors were able to identify a rare hematopoietic progenitor population that most closely resembles fetal liver HSC/multipotent progenitors (MPPs). This population was rare in the culture, and its frequency declined over time in culture. The transcriptional differences identified between in vivo– and in vitro–generated HSC/MPPs might help explain the functional deficiencies of hPSCs-derived blood derivatives to engraft and reconstitute hematopoiesis in vivo and could ultimately be exploited to improve their production in vitro and their therapeutic potential. For example, EGR1 and other members of the early response genes family (ZFP36L1, NR4A1, FOS, JUN, and JUNB) were expressed at a lower level in the in vitro–produced HSC/MPP, and so these genes could be used in direct programming strategies to manufacture HSCs. Stem cell–based applications rely on the ability to activate several endogenous genes simultaneously to modify cell fate. Recent developments of multiplexed genetic intervention of hPSCs using catalytically dead Cas9 (dCas9) activator systems represent an unprecedented tool to simultaneously activate several endogenous genes for prospective generation of functional HSCs.9
The limitations in the production of reconstituting HSCs in vitro from human hPSCs could be explained by the incapacity either to generate or to maintain HSCs. It is possible that the culture conditions employed to differentiate hPSCs also promote the differentiation of HSCs into lineage-restricted progenitors. In a recent study, hPSCs were differentiated until the EHT stage, when they were genetically manipulated to overexpressed transcription factors. The transgenes were induced when the cells were transferred into the bone marrow to support the EHT process, and this resulted in the reconstitution of the hematopoietic system.10 This underlines the importance of the microenvironment and implies that functional HSCs can be generated from hPSCs, suggesting that in vitro culture conditions are unable to support their maintenance. Further studies are required to assess the functional properties of the HSC-like population identified by Fidanza et al and to test if appropriate culture of this population could help to define the limitation of HSCs production. The methology developed by Fidanza et al represents a novel approach that can be now applied to any model of in vitro differentiation, thus helping to overcome the challenges in the production of other therapeutic cell types from hPSCs.
Conflict-of-interest disclosure: The authors declare no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal