TO THE EDITOR:
Since December 2019, a novel human coronavirus, SARS-CoV-2, has emerged from China where the first cases of COVID-19 were described.1,2 It is the third highly pathogenic coronavirus introduced in humans from animal reservoirs3,4 and has spread worldwide, leading to an unprecedented pandemic. By August 2020, more than 20 million cases have been reported worldwide, including almost 0.8 million deaths. France has reported more than 200 000 cases and 30 000 deaths with an epidemic peak at week 14 (30 March-5 April).5 SARS-CoV-2 is mostly transmitted through airborne droplets,6-8 with a reproduction number (R0) varying between 2.5 and 3.5 before the implementation of control measures.9-11 COVID-19 incubation is short (5.7-5.9 days)12 and mostly asymptomatic or with moderate symptoms, but 20% to 25% of infected individuals develop severe symptoms, some of them needing intensive care.6,13,14 First reported data suggested the presence of SARS-CoV-2 RNAemia in patients with severe symptoms,10,11,15 but not in infected asymptomatic individuals.16,17 However, a Chinese study described 4 of more than 7400 blood donors who had extremely low plasma viral load: 2 remaining asymptomatic and 2 declaring fever after donation.18 Therefore, the question of transmission of SARS-CoV-2 through transfusion became relevant. In the context of the SARS-CoV-2 pandemic, we relied on the French hemovigilance network to investigate the presence of SARS-CoV-2 RNA in the plasma of blood donors reporting COVID-19–like symptoms after donation or in donations involved in recipient-initiated trace-back studies, whenever a patient develop symptoms related to COVID-19 shortly after a blood transfusion. This investigation was conducted in accordance with the Declaration of Helsinki. We also addressed the issue of the infectivity of positive samples using direct and enriched virus culture.
From March 2020, blood donors were asked to report to the French National Blood Service (EFS) any SARS-CoV-2–positive reverse transcriptase-polymerase chain reaction (RT-PCR) result or COVID-19–like symptoms occurring within 15 days after donation (postdonation information [PDI]). Moreover, whenever a patient developed COVID-19–like symptoms within 14 days after a blood transfusion, involved donations were traced back. For each donation meeting 1 of these criteria, the cryopreserved plasma sample from the EFS repository biobank was initially tested for SARS-CoV-2 RNA using SARS-CoV-2 RNA R-GENE assay (BioMérieux, Craponne, France) and then with a SARS-CoV-2 real-time PCR (NRC-rtPCR) set up by the National Reference Center for respiratory viruses (Pasteur Institute).19 Positive samples were sequenced after amplification by an in-house seminested RT-PCR targeting the RdRp region (Table 1) and tested for total anti–SARS-CoV-2 antibodies using Platelia SARS-CoV-2 Total Ab (Bio-Rad, Marnes-la-Coquette, France). SARS-CoV-2 infectivity was evaluated in plasma retrieved from the plasma units using virus isolation on Vero E6 cells (2 successive 6-day cultures). To improve sensitivity, virions were concentrated by ultracentrifugation from 12 mL plasma or 10.5 mL plasma on iodixanol pad for RT-PCR and viral culture, respectively.
Results of virologic investigations of the 3 SARS-CoV-2–positive blood donations
| No. of donation . | Detection by rtPCR on neat plasma, n positive/n tested (Ct) . | Detection by rtPCR after UC, n positive/n tested (Ct) . | RT-PCR‡/sequencing . | Virus culture . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R-GENE PCR1* . | R-GENE PCR2* . | NRC rtPCR† . | R-GENE PCR1 . | R-GENE PCR2 . | ||||||||
| NC . | RdPd . | E . | RdPd target 1 . | RdPd target 2 . | NC . | RdPd . | E . | Plasma . | After UC . | Plasma . | After i-UC . | |
| 1 | POS 1/1 (37.9) | NEG 0/1 | NEG 0/1 | NEG 0/4 | Pos 2/4 (37.0; 37.1) | POS 1/1 (35.3) | POS 1/1 (41.6) | NEG O/1 | NEG | POS 1/1 | NEG | NEG |
| 2 | POS 1/1 (37.5) | NEG 0/1 | NEG 0/1 | POS 1/3 (39.3) | POS 1/3 (37.4) | POS 1/2 (37.0) | NEG 0/2 | NEG 0/1 | NEG | POS 1/2 | NEG | NEG |
| 3 | POS 1/1 (38.0) | NEG 0/1 | POS 1/1 (38.4) | NEG 0/2 | NEG 0/2 | POS 1/2 (38.3) | NEG 0/2 | POS 1/1 (42.2) | NEG | NEG 0/2 | NEG | NEG |
| No. of donation . | Detection by rtPCR on neat plasma, n positive/n tested (Ct) . | Detection by rtPCR after UC, n positive/n tested (Ct) . | RT-PCR‡/sequencing . | Virus culture . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R-GENE PCR1* . | R-GENE PCR2* . | NRC rtPCR† . | R-GENE PCR1 . | R-GENE PCR2 . | ||||||||
| NC . | RdPd . | E . | RdPd target 1 . | RdPd target 2 . | NC . | RdPd . | E . | Plasma . | After UC . | Plasma . | After i-UC . | |
| 1 | POS 1/1 (37.9) | NEG 0/1 | NEG 0/1 | NEG 0/4 | Pos 2/4 (37.0; 37.1) | POS 1/1 (35.3) | POS 1/1 (41.6) | NEG O/1 | NEG | POS 1/1 | NEG | NEG |
| 2 | POS 1/1 (37.5) | NEG 0/1 | NEG 0/1 | POS 1/3 (39.3) | POS 1/3 (37.4) | POS 1/2 (37.0) | NEG 0/2 | NEG 0/1 | NEG | POS 1/2 | NEG | NEG |
| 3 | POS 1/1 (38.0) | NEG 0/1 | POS 1/1 (38.4) | NEG 0/2 | NEG 0/2 | POS 1/2 (38.3) | NEG 0/2 | POS 1/1 (42.2) | NEG | NEG 0/2 | NEG | NEG |
E, envelope protein; Ct, cycle threshold; i-UC, ultracentrifugation on iodixanol pad (10.5 mL plasma); NEG, negative; NC, nucleocapsid gene; POS, positive; RdPd, polymerase gene; UC, ultracentrifugation (12 mL plasma).
SARS-COV-2 R-GENE assay (BioMérieux).
rtPCR from the National Reference Center for respiratory viruses (NCR rt-PCR, Pasteur Institute).
RT-PCR primers: 5′-GCTTTTCAAACTGTCAAACC-3′ and 5′-GGTGAGGGTTTTCTACATCA-3′; hemi-nested PCR primers: 5′-TGCTGTGTCTAAGGGTTTCT-3′ and 5′-GGTGAGGGTTTTCTACATCA-3′.
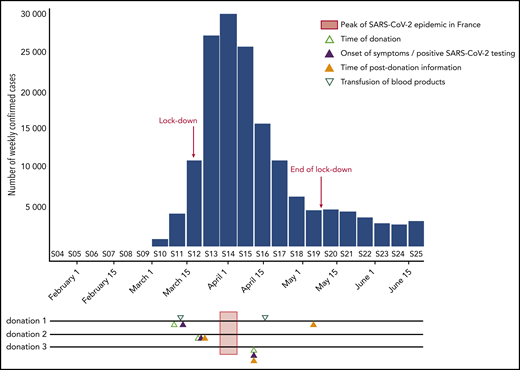
From 20 January to 29 May, 311 donations were investigated including 268 PDI and 43 trace-back donations. Three of 268 PDI donations (1.1%) tested positive for SARS-CoV-2 RNA with high Ct values (Ct > 37). They all tested negative for anti–SARS-CoV-2 antibodies. Detailed results relative to these 3 donations are given in Table 1 and Figure 1. Briefly, the first donor donated whole blood on week 11. She tested positive for SARS-CoV-2 in a nasal swab at day 4 after donation but remained asymptomatic and reported PDI at D56. Three products were prepared from the blood donation. The pathogen-reduced platelet concentrate was transfused at day 3 after donation to a recipient who remained asymptomatic. The red blood cells were transfused to a COVID-19 patient at day 37 after donation, and the plasma unit was not transfused. The plasma tested positive with R-GENE PCR1 and NRC-rtPCR and with in-house RT-PCR after ultracentrifugation. The second donor donated whole blood on week 12, experienced fever, cephalgia, anosmia, and ageusia at day 2, and reported PDI at day 4. She was not tested for SARS-CoV-2, and no blood product from her donation was transfused. The plasma tested positive with R-GENE PCR1 and NRC-rtPCR and the in-house RT-PCR after ultracentrifugation. The last donor reported PDI after noticing ageusia and anosmia on the same day as blood donation. No test for COVID-19 was performed, and no blood product from her donation was transfused. The plasma was positive with both R-GENE PCR1 and PCR2. No amplification by seminested RT-PCR was obtained, even after virus concentration. Virus isolation failed on neat and ultracentrifuged plasma from the 3 donations. Last, the partial 535-nt sequences obtained from donations 1 and 2 (MT708232 and MT708233, respectively) were identical to SARS-CoV2 Wuhan-Hu-1 isolate (MN908947) and 62 worldwide sequences used as references.
Timeline SARS-CoV-2RNA–positive donations with regard to the epidemic curve in France. The weekly number of incident confirmed COVID-19 cases, as reported to the French National Public Health Agency (SpF), is shown by the blue bars. On the timelines corresponding to the 3 SARS-CoV-2–positive donations are shown the time of donation (open light green triangle), the onsets of symptoms or the day of positive testing (full blue triangle), the time of transfusion of blood products (open green triangle), and the time of postdonation information (full orange triangle). The epidemic peak (week 14) is indicated by a light red bar crossing the timelines.
Timeline SARS-CoV-2RNA–positive donations with regard to the epidemic curve in France. The weekly number of incident confirmed COVID-19 cases, as reported to the French National Public Health Agency (SpF), is shown by the blue bars. On the timelines corresponding to the 3 SARS-CoV-2–positive donations are shown the time of donation (open light green triangle), the onsets of symptoms or the day of positive testing (full blue triangle), the time of transfusion of blood products (open green triangle), and the time of postdonation information (full orange triangle). The epidemic peak (week 14) is indicated by a light red bar crossing the timelines.
In addition, 4 blood immunocompromised recipients from 5 to 67 years of age were involved in traceback. They received between 2 and 25 blood products, including 18 red blood cell units and 23 pathogen-reduced platelets units. None of the 43 traced-back repository samples tested SARS-CoV-2 RNA positive, in particular, those corresponding to the 18 red blood units, which were not pathogen reduced.
Although the data on SARS-CoV suggested that human sarbecoviruses were not transmissible through transfusion,15,16 the presence of SARS-CoV-2 RNA in plasma of asymptomatic donors may suggest a risk for blood safety.18 Thus far, no studies reported the infectivity of SARS-CoV-2 RNA–positive plasma from asymptomatic individuals. We found that a very small fraction of donors, even remaining asymptomatic after donation, can be RNAemic for SARS-CoV-2. Although the prevalence of this RNAemia cannot be estimated from our study, we can assume that less than 1% of donors infected by SARS-CoV-2 at the time of donation exhibit a viremia, with low viral loads, as suggested by high Ct, and the random positivity observed in replicates. Infectivity of positive plasma was not evidenced in cell culture experiments, despite a concentration step, and recipient-initiated traceback yielded no positive donation. Altogether, although blood products contain a great volume of residual plasma (approximately 20 mL) that may increase the global amount of transmitted virus, our data support the absence of proven SARS-CoV-2 transmission by transfusion thus far. This may be because of low viral loads, but the presence of defective viruses, fragmented RNA, or a genuine RNAemia without intact viral particles cannot be ruled out. The pathogen-reduction treatment of platelet units may also explain the absence of transmission by platelet concentrates. In addition, the absence of anti–SARS-CoV-2 antibodies in the 3 positive samples rule out that the absence of transmission is because of the neutralizing potential of the plasma.
Based on the assumption that viremia could lead to transfusion transmission, the minimum infectious dose through blood, if any, is unknown for SARS-CoV-2. Nevertheless, it might be probably higher than RNA levels found in asymptomatic blood donors, given that blood is not the natural route of infection for respiratory viruses. Interestingly, the 4 recipients involved in recipient-initiated traceback studies were immunocompromised, and the minimum infectious dose might be lower than in immunocompetent patients. Last, our work also highlights that, during an epidemic, blood products may be transfused to individuals already infected, making it difficult to distinguish between interindividual and transfusion transmission.
In conclusion, although SARS-CoV-2 transfusion transmission risk cannot be totally excluded, we demonstrated that viremia was extremely rare in asymptomatic blood donors, viral RNA levels were very low when detected, and the corresponding plasma was not infectious in cell culture. The presence in the plasma of nucleic acid related to emerging viruses for which transmission by blood is not the natural mode of contamination does not necessarily imply a threat to blood safety. Proving transfusion transmission is extremely difficult in real time, and hemovigilance is therefore a major corner stone of blood safety.
Acknowledgments
The authors thank all medical doctors and medical staff involved in hemovigilance and the staff working at the EFS biobank.
Authorship
Contribution: P.C., S.L., J.P., P.T., and P.M. conceived the study; P.C., D.C., V.S., and V.E. implemented the methods; Q.L., L.B., and J.G. performed the experiments; L.C., P.T., and P.M. organized and supervised the collection of hemovigilance data; and all authors read and approved the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Pierre Cappy, INTS-DATS, 6 rue Alexandre Cabanel, 75015 Paris, France; e-mail: pcappy@ints.fr.