Key Points
RNA-based Ig repertoire sequencing (RACE-RepSeq) is a highly sensitive approach to detect low-abundance POEMS syndrome-related clones.
RACE-RepSeq with full-length Ig light chain variable domain sequencing reveals specific mutational patterns in POEMS syndrome.
Abstract
Polyneuropathy, organomegaly, endocrinopathy, monoclonal gammopathy, and skin changes (POEMS) syndrome is a rare multisystem disease resulting from an underlying plasma cell (PC) dyscrasia. The pathophysiology of the disease remains unclear, but the role of the monoclonal immunoglobulin (Ig) light chain (LC) is strongly suspected because of the highly restrictive usage of 2 λ variable (V) domains (IGLV1-40 and IGLV1-44) and the general improvement of clinical manifestations after PC clone-targeted treatment. However, the diagnostic value of Ig LC sequencing, especially in the case of incomplete forms of the disease, remains to be determined. Using a sensitive high-throughput Ig repertoire sequencing on RNA (rapid amplification of cDNA ends-based repertoire sequencing [RACE-RepSeq]), we detected a λ LC monoclonal expansion in the bone marrow (BM) of 83% of patients with POEMS syndrome, including some in whom BM tests routinely performed to diagnose plasma cell dyscrasia failed to detect λ+ monoclonal PCs. Twenty-four (83%) of the 29 LC clonal sequences found were derived from the IGLV1-40 and IGLV1-44 germline genes, as well as 2 from the closely related IGLV1-36 gene, and all were associated with an IGLJ3*02 junction (J) gene, confirming the high restriction of VJ region usage in POEMS syndrome. RACE-RepSeq VJ full-length sequencing additionally revealed original mutational patterns, the strong specificity of which might crucially help establish or eliminate the diagnosis of POEMS syndrome in uncertain cases. Thus, RACE-RepSeq appears as a sensitive, rapid, and specific tool to detect low-abundance PC clones in BM and assign them to POEMS syndrome, with all the consequences for therapeutic options.
Introduction
Polyneuropathy, organomegaly, endocrinopathy, monoclonal gammopathy, and skin changes (POEMS) syndrome is a multisystem disorder. Apart from the above manifestations, sclerotic bone lesions, edema, anasarca, thrombocytosis, and various other symptoms are common. Diagnosis relies on the presence of at least 3 major criteria and 1 minor criteria.1 Among major criteria, monoclonal plasma cell (PC) disorder and polyradiculoneuropathy are mandatory. Other major criteria include bone lesions, high level of vascular endothelial growth factor (VEGF), and Castleman disease. Minor criteria are numerous, reflecting the heterogeneity of the disease manifestations; they include organomegaly, endocrinopathy, skin changes, papilledema, extravascular volume overload, and thrombocytosis.
POEMS syndrome has been recently incorporated within the spectrum of the monoclonal gammopathies of clinical significance.2 Indeed, even if the pathophysiology of the disease is yet unknown, an efficient treatment of the underlying PC clone usually results in improvement of all manifestations. The clones are often small,3,4 and almost always characterized by the production of a λ light chain (LC) deriving from any of 2 variable (V) germline genes, IGLV1-40 or IGLV1-44.5,6 As a consequence, it has been suggested that, through still-unknown pathways, the monoclonal LC structure might specifically trigger the major biologic anomaly suspected to account for the POEMS pathophysiology: VEGF hyperproduction. High VEGF levels indeed could appear to account for most features of the disease,2,7 despite the ambiguous efficacy of anti-VEGF treatment.8 Immunochemically, a small free λ LC component is virtually always associated with a complete immunoglobulin, most frequently immunoglobulin Aλ (IgAλ), or IgGλ. Rarely, the λ LC is the sole monoclonal component. Despite the fact there is usually a slight increase in serum λ free LC level, nearly 80% of the cases present with a normal κ/λ ratio.9,10 Treatment is chosen depending on the characteristics of the underlying PC clone. Radiotherapy is used for localized plasmacytoma, whereas systemic chemotherapy is required in patients with bone marrow (BM) infiltration or with too many or no bone lesions.1,11 Thus, detection and identification of a BM PC clone is a crucial step of management. To date, a PC clone is found in up to 66% of patients with POEMS syndrome, using flow cytometry, immunohistochemistry, and/or in situ hybridization technics.12 However, this percentage may be either underestimated because of a low level PC infiltration, especially in the presence of a concomitant plasmacytoma,13 or overestimated when nonrelevant clones are present in the BM. In this setting, because of the homogeneity of Ig V domain usage,5,6 Ig sequencing may be of great value to determine the pathogenicity of a detected clone, particularly in patients with atypical clinical presentation.
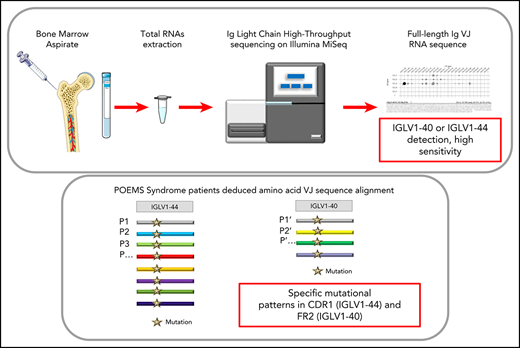
Next-generation sequencing (NGS) has changed the paradigm of Ig repertoire analysis and detection of minimal residual disease in B-cell malignancies, including multiple myeloma.14 Kawajiri-Manako et al showed the possibility of using NGS to identify PC clone LC rearrangements on DNA extracted from the BM of patients with POEMS syndrome.13 However, because of the low clonal expansion of PCs in BM, they detected clonal λ LC rearrangements in only 11 of 30 patients. Moreover, the PCR amplification design usually used for characterizing the Ig repertoire on DNA only encompasses the CDR3 region. Although this strategy might give a good evaluation of IgH VDJ genes, it can be expected with a poor efficacy when evaluating the clonality of light chain genes (which carry little or absent N insertions at the VJ region, and which usually display more canonical rearrangements than VHDJH genes with poorly diversified CDR3 length). This strategy of characterizing clones through a short CDR3 stretch instead of the full VJ regions also overlooks most of the clonal diversification related to somatic hypermutation (thus masking intraclonal diversity), and might even be unable to assign a VJ rearrangement to one or the other member of a VL subgroup (notably in the case of the closely related V germline genes IGLV1-44 and IGLV1-36).13 In contrast, we propose here a method for high-resolution characterization of the full-length VJ region. This relies on a sensitive and rapid cDNA repertoire analysis of BM or bone lesions/plasmacytoma through high-throughput sequencing. We evaluate here this NGS approach to detect and characterize λ LC clones in POEMS syndrome to secure the diagnosis and management of this disease.15
Methods
Patient protections
This study has received approval for retention and treatment of biological samples from the Comité de Protection des Personnes (CPP DC-2008-111), and Commission Nationale de l'Informatique et des Libertés (CNIL DR211392) and Comité Consultatif sur le Traitement de l'Information en Matière de Recherche dans le Domaine de la Santé (CCTIRS N°1158) approvals for additional data processing. Research was conducted in accordance with the Declaration of Helsinki
RNA sample preparation
Total RNAs were extracted and prepared from fresh BM aspirates or plasmacytoma biopsies, using TRIzol reagent (Ambion). When available, we preferentially used Tempus Blood RNA Tubes (Applied Biosystems) or the equivalent to collect BM aspirates and RNA later (Invitrogen) for plasmacytoma biopsies. This ensures a high RNA quality without any other treatments.
TOPO cloning and sequencing
Complementary DNAs were obtained by reverse transcription performed using the High Capacity cDNA Archive Kit (Applied Biosystems) and 1 µg total RNAs. Polymerase chain reaction (PCR) amplification was performed using a 3′ consensus primer complementary to all λ constant regions (5′ CTCCCGGGTAGAAGTCACT 3′) and a 5′ consensus primer for all Vλ subgroups leader regions (5′ ATGGCCTGGDYYVYDCTVYTYCT 3′). PCR products were then cloned into pCR2.1 TOPO vector (Invitrogen), and DNA sequencing was carried out using Big-Dye Terminators (Applied Biosystems) on a 16-capillaries electrophoresis system 3130 XL (Applied Biosystems). Sequences were analyzed using FinchTV software and aligned with MultAlin (http://multalin.toulouse.inra.fr/multalin/), and the λ VJ rearrangement was determined using the IMGT/V-QUEST tool (http://imgt.org/).16-18
High-throughput sequencing on Illumina MiSeq sequencer
The protocol was adapted from previous studies using high-throughput sequencing on cDNA to analyze BCR repertoire.19,20 For each sample, up to 2 µg total RNAs were used for high-throughput sequencing.
5′ Rapid amplification of cDNA ends PCR
First, a mix containing up to 2 µg RNAs, 1 µL consensus reverse primer of all λ constant regions (5′CTGGCCGCYTACTTGTTGTT 3′), and 1 µL 2′-deoxynucleoside 5'-triphosphate solution mix (New England Biolabs), adjusted to a final volume of 12 µL with water, was incubated for 3 minutes at 72°C and 2 minutes at 42°C. After a short spin, the mix was placed on ice for 2 minutes. Then, 4 µL ProtoScript buffer, 2 µL dithiothreitol solution, 1 µL ProtoScriptII (New England Biolabs), and finally 1 µL cap-race primer (5′ AAGCAGTGGTATCAACGCAGAGTACAT[GGGG] 3′, with the 4 G between brackets being ribonucleotides) was added. This mix was incubated 90 minutes at 42°C and 10 minutes at 70°C, and then stored at 4°C. At the end of the reaction, 20 µL RNAse free water was added
Library preparation
The λ cDNA was amplified with Taq Phusion (New England Biolabs), using a universal forward universal primer mix (5′ CTAATACGACTCACTATAGGGC 3′ and 5′ CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT 3′, in a ratio of 4:1), as described,21 and a reverse primer hybridizing within the λ constant exons (5′ GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGTTGGCTTGYAGCTCCTCAG 3′). Four PCR replicates of 25 µL were performed. Cycling conditions were 30 seconds at 98°C, 32 cycles of 30 seconds at 98°C, 30 seconds at 65°C, 30 seconds at 72°C, and final elongation 5 minutes at 72°C. The 4 replicates were pooled and migrated in a 2% agarose gel (alternatively, Ampure XP beads can be used). The band of interest (±550 pb) was cut and the DNA eluted in 30 µL elution buffer, using the kit Nucleospin gel and PCR clean-up (Macherey-Nagel). Then, Illumina sequencing adapter and tag sequences were added by primer extension. For this, previous DNA was reamplified with Taq Phusion. Four PCR replicates of 25 µL were made using primers containing a unique couple of tags per sample (a tag is a unique combination of 8 nucleotides). Cycling conditions were 30 seconds at 98°C, only 12 cycles of 30 seconds at 98°C, 30 seconds at 65°C, 30 seconds at 72°C, and final elongation 5 minutes at 72°C. The 4 replicates were mixed, and another migration in a 2% agarose gel was performed (alternatively, Ampure XP beads can also be used). The band of interest (±600 pb) was cut and DNAs were eluted twice in 30 µL elution buffer, using the kit Nucleospin gel and PCR clean-up (macherey-Nagel).
Sequencing on Illumina MiSeq and repertoire analysis
Resulting amplicons were sequenced on an Illumina MiSeq sequencing system using MiSeq Reagent Kit V3 600 cycles (alternatively, MiSeq reagent kit V2 can be used, but with lower cluster density and shorter sequences). Paired reads were merged using FLASH.22 Repertoire analysis was performed using the VIDJIL tool (http://vidjil.org/)23 (supplemental Figure 1, available on the Blood Web site) and IMGT/HIGHV-QUEST tool (http://imgt.org/), resulting in consensus sequences for each clonotype.21,24 Using MiSeq Reagent Kit V3, the sequencing of 24 samples allowed us to obtain an average of 325 000 reads per sample (from 95 500 to 554 516 per sample). Data are available at European Nucleotide Archive (ENA) database under accession number PRJEB35157. Fourteen VJ sequences obtained both by Sanger and MiSeq methods were compared and found to be identical, proving the accuracy of the MiSeq sequencing for Ig repertoire (examples in supplemental Figure 2). Five samples (p9, p22, p23, p29, p30) were analyzed twice with both a V2 and V3 Illumina kit to demonstrate the reproducibility of the MiSeq sequencing method (supplemental Figure 2).
Sequencing of IGLV genes on DNA
DNA extraction from a lymph node biopsy and sequencing of the IGLV gene were performed using the EuroClonality/BIOMED-2 guidelines as previously described,25,26 with an IGLV forward consensus primer (5′ ATTCTCTGGCTCCAAGTCTGGC 3′) specific to the FR3 region and an IGLJ reverse consensus primer (5′ CTAGGACGGTGAGCTTGGTCCC 3′).
Results
A total of 35 samples from patients with confirmed POEMS syndrome were retrospectively analyzed by high-throughput sequencing (rapid amplification of cDNA ends-based repertoire sequencing [RACE-RepSeq]), except for 4 patients that were characterized only with TOPO cloning and Sanger sequencing (p1, p3, p6, p21). Twenty-four patients (69%) had bone lesions and 15 patients (43%) had BM monoclonal plasma cell infiltration identified by cytology and/or flow cytometry (usually non-Next Generation Flow) on BM aspirates or immunohistology on BM biopsies (Table 1). BM samples were available in 32 patients. The remaining samples were obtained from bone lesions in 3 cases, (p7, p8, p18). In 1 patient, biopsy samples both from BM and from a bone lesion were analyzed (p15). In another patient with an atypical form of POEMS syndrome (p33), both biopsy samples from BM and those from a lymph node were analyzed. Ig RNA sequencing allowed finding an IGLV clone in all patients with a detectable BM PC clone (Table 1). Interestingly, a clonal IGLV gene was found in 13 of 19 BM samples with negative cytology and cytometry or with monotypic κ PCs (Table 1). All but 1 of these patients presented with 1 or several plasmacytomas. A comparison of the TOPO cloning method with our RACE-RepSeq method revealed that 11 of 16 samples previously negative for IGLV clone using Sanger sequencing were subsequently found positive by NGS. Overall, we detected at least 1 IGLV clone in 30 (86%) of 35 patients and in 26 (81%) of 32 patients whose BM samples were analyzed. For patient 15, the same monoclonal LC sequences were detected in both BM and bone lesion samples (supplemental Figure 1). Concerning patient 33, with an atypical form of POEMS syndrome (polyneuropathy, circulating monoclonal IgGλ, elevated serum VEGF, but no bone lesion or monoclonal plasmocytosis in the BM and multiple lymph nodes with an aspect of follicular hyperplasia with monoclonal λ+ PC), the BM was negative by RACE-RepSeq, but we detected an IGLV monoclonal sequence in a lymph node biopsy (included in paraffin) obtained from a Sanger DNA sequencing surrounding the V-J junction (CDR3) only.
Clinical, biological and monoclonal λ LC characteristics of POEMS syndrome patients
| Patient ID . | BM clonal plasmocytosis . | % BM plasma cells (abnormal: yes/no) . | BM typing flow cytometry . | BM biopsy . | Bone lesions . | SANGER analysis clone, yes/no . | NGS analysis clone, yes/no . | IGLV gene . | IGLJ gene . | Mutation . | Monoclonal Ig (electrophoresis +immunofixation) . | λFLC, mg/L (range, 5.70-26.3) . | kFLC/λFLC ratio (range, 0.26-1.65) . | VEGF in pg/mL (Nl <500 pg/L) . | IGVH gene or other IGLV clone . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BM sequencing | |||||||||||||||
| p4 | Negative | 2 (no) | No clone | ND | Yes | Yes | Yes | IGLV1-44 | IGLJ3*02 | 38→P | αλ | 15.8 | 1 | 1618 | IGHV3-11 and IGHV4-39 |
| p6 | Negative | 0.7 (no) | No clone | No clone | Yes | Yes | ND | IGLV1-44 | IGLJ3*02 | 38→P | ɣλ | 10.8 | 1.72 | 420 | |
| p9 | Negative | 1.5 (no) | No clone | ND | Yes | No | Yes | IGLV1-44 | IGLJ3*02 | 38→P | ɣλ | 23.3 | 0.69 | 162 | Other clone (IGLV3-1/IGLJ2*01) |
| p10 | Negative | 2 (no) | No clone | ND | Yes | Yes | Yes | IGLV1-44 | IGLJ3*02 | 38→P | αλ | 5.78 | 1.08 | 430 | IGHV3-9 |
| p11 | Negative | 2.5 (no) | No clone | ND | Yes | No | Yes | IGLV1-44 | IGLJ3*02 | 38→P | ɣλ | 100 | 0.75 | 3752 | |
| p13 | Negative | 2.5 (no) | No clone | ND | Yes | No | Yes | IGLV1-44 | IGLJ3*02 | 38→P | αλ | 161 | 0.59 | 63 332 | |
| p14 | Negative | 1.3 (no) | ND | No clone | Yes | No | Yes | IGLV1-44 | IGLJ3*02 | 38→P | αλ | 32 | 0.74 | 2300 | |
| p17 | Negative | <5 (no) | No clone | No clone | No | Yes | Yes | IGLV1-44 | IGLJ3*02 | 38→N | αλ | — | — | 2000 | |
| p1 | Positive (λ) | 0.5 (yes) | λ clone | λ clone | Yes | Yes | ND | IGLV1-44 | IGLJ3*02 | 38→P | αλ | 26.3 | 0.53 | 1965 | IGHV4-38-2 |
| p2 | Positive (λ) | 13 (yes) | λ clone | ND | Yes | Yes | Yes | IGLV1-44 | IGLJ3*02 | 38→P | ɣλ | 104 | 0.13 | 1082 | IGHV2-26 |
| p3 | Positive (λ) | 9 (yes) | λ clone | ND | Yes | Yes | ND | IGLV1-44 | IGLJ3*02 | 38→P | αλ | 35.4 | 0.38 | 7000 | |
| p5 | Positive (λ) | 3.5 (no) | λ clone | ND | No | No | Yes | IGLV1-44 | IGLJ3*02 | 38→P | αλ | 217 | 0.06 | 2725 | |
| p12 | Positive (PND) | 35 (yes) | ND | ND | No | No | Yes | IGLV1-44 | IGLJ3*02 | 38→S | αλ | 57.5 | 0.82 | 7800 | |
| p15 | Positive (λ) | 6 (yes) | λ clone | ND | Yes | Yes | Yes† | IGLV1-44 | IGLJ3*02 | 38→A | αλ | 72 | 0.21 | 9055 | |
| p16 | Positive (λ) | 5 (unknown) | λ clone | ND | No | Yes | Yes | IGLV1-44 | IGLJ3*02 | 38→A | ɣλ | 51.9 | 0.25 | 739 | |
| p23 | Negative | <5 (no) | No clone | No clone | Yes | No | Yes | IGLV1-40 | IGLJ3*02 | 40→N | No monoclonal Ig | 46 | 0.76 | 845 | |
| p24 | Negative | 4 (no) | ND | ND | Yes | Yes | Yes | IGLV1-40 | IGLJ3*02 | 40→N | — | 47 | 0.5 | 1300 | |
| p20 | Positive (k) | 2.3 (yes) | κ clone | ND | Yes | No | Yes | IGLV1-40 | IGLJ3*02 | 40→N | ɣλ and ɣk | 150 | 0.55 | 1400 | |
| p19 | Positive (PND) | 2 (yes) | λ clone | ND | No | Yes | Yes | IGLV1-40 | IGLJ3*02 | 40→N | αλ | 43.3 | 0.61 | 11 188 | |
| p21 | Positive (PND) | 22 (yes) | ND | ND | Yes | Yes | ND | IGLV1-40 | IGLJ3*02 or 01 or IGLJ2*01 | 40→Q | ɣλ | 134 | 0.09 | 1423 | |
| p22 | Positive (λ) | 3 (yes) | λ clone | ND | No | Yes | Yes | IGLV1-40 | IGLJ3*02 | 40→N | αλ | 32 | 0.74 | 7290 | IGHV2-70 |
| p25 | Positive (λ) | 0 (no) | λ clone | No clone | No | Yes | Yes | IGLV1-36 | IGLJ3*02 | — | ɣλ | 37.8 | 0.72 | 5000 | Other clone (IGLV2-14/IGLJ3*02) |
| p26 | Positive (λ) | 1 (no) | ND | λ | No | Yes | Yes | IGLV1-36 | IGLJ3*02 | — | No monoclonal Ig | 107 | 0.36 | 7550 | |
| p29 | Negative | 1% (no) | No clone | No clone | Yes | No | Yes | IGLV3-1 | IGLJ2*01 | — | ɣλ | 40.2 | 0.75 | 3275 | |
| p30 | Negative | <5 (no) | ND | No clone | No | No | Yes | IGLV3-25 | IGLJ1*01 | — | ɣλ | — | — | 1966 | |
| p27 | Positive (λ) | 2 (no) | λ clone | ND | Yes | No | Yes | IGLV3-1 | IGLJ3*01 | — | ɣλ | 35 | 0.45 | 1210 | |
| p31 | Negative | 1%-2.5% (no) | No clone | ND | Yes | No | No | No clone | No clone | — | No monoclonal Ig | 41 | 0.69 | 1700 | |
| p32 | Negative | 2 (no) | ND | ND | Yes | No | No | No clone | No clone | — | ɣλ | 51.3 | 0.46 | 1190 | |
| p33 | Negative | 3.5 (no) | No clone | ND | No | No | No‡ | No clone‡ | No clone‡ | — | ɣλ | 25.2 | 1.31 | 1146 | |
| p34 | Negative | 0.3 (no) | No clone | ND | Yes | No | No | No clone | No clone | — | ɣλ | 21.8 | 0.54 | 705 | |
| p35 | Negative | 2 (no) | No clone | ND | Yes | ND | No | No clone | No clone | — | αλ | — | — | 2550 | |
| p36 | Negative | 2 (no) | ND | ND | No | No | No | No clone | No clone | — | αλ | 27 | 1.33 | 665 | |
| Bone lesion sequencing | |||||||||||||||
| p7 | Negative | 2 (no) | ND | ND | Yes | Yes | Yes | IGLV1-44 | IGLJ3*02 | 38→A | ɣλ | 15.3 | 0.77 | 1419 | |
| p8 | Negative | 1.5 (no) | No clone | ND | Yes | Yes | Yes | IGLV1-44 | IGLJ3*02 | 38→A | ɣλ and αk | 55.6 | 0.99 | 3560 | |
| p18 | Positive (k) | ND | ND | κ | Yes | Yes | Yes | IGLV1-44 | IGLJ3*02 | 38→P | ɣk and ɣλ | 21.6 | — | 1100 |
| Patient ID . | BM clonal plasmocytosis . | % BM plasma cells (abnormal: yes/no) . | BM typing flow cytometry . | BM biopsy . | Bone lesions . | SANGER analysis clone, yes/no . | NGS analysis clone, yes/no . | IGLV gene . | IGLJ gene . | Mutation . | Monoclonal Ig (electrophoresis +immunofixation) . | λFLC, mg/L (range, 5.70-26.3) . | kFLC/λFLC ratio (range, 0.26-1.65) . | VEGF in pg/mL (Nl <500 pg/L) . | IGVH gene or other IGLV clone . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BM sequencing | |||||||||||||||
| p4 | Negative | 2 (no) | No clone | ND | Yes | Yes | Yes | IGLV1-44 | IGLJ3*02 | 38→P | αλ | 15.8 | 1 | 1618 | IGHV3-11 and IGHV4-39 |
| p6 | Negative | 0.7 (no) | No clone | No clone | Yes | Yes | ND | IGLV1-44 | IGLJ3*02 | 38→P | ɣλ | 10.8 | 1.72 | 420 | |
| p9 | Negative | 1.5 (no) | No clone | ND | Yes | No | Yes | IGLV1-44 | IGLJ3*02 | 38→P | ɣλ | 23.3 | 0.69 | 162 | Other clone (IGLV3-1/IGLJ2*01) |
| p10 | Negative | 2 (no) | No clone | ND | Yes | Yes | Yes | IGLV1-44 | IGLJ3*02 | 38→P | αλ | 5.78 | 1.08 | 430 | IGHV3-9 |
| p11 | Negative | 2.5 (no) | No clone | ND | Yes | No | Yes | IGLV1-44 | IGLJ3*02 | 38→P | ɣλ | 100 | 0.75 | 3752 | |
| p13 | Negative | 2.5 (no) | No clone | ND | Yes | No | Yes | IGLV1-44 | IGLJ3*02 | 38→P | αλ | 161 | 0.59 | 63 332 | |
| p14 | Negative | 1.3 (no) | ND | No clone | Yes | No | Yes | IGLV1-44 | IGLJ3*02 | 38→P | αλ | 32 | 0.74 | 2300 | |
| p17 | Negative | <5 (no) | No clone | No clone | No | Yes | Yes | IGLV1-44 | IGLJ3*02 | 38→N | αλ | — | — | 2000 | |
| p1 | Positive (λ) | 0.5 (yes) | λ clone | λ clone | Yes | Yes | ND | IGLV1-44 | IGLJ3*02 | 38→P | αλ | 26.3 | 0.53 | 1965 | IGHV4-38-2 |
| p2 | Positive (λ) | 13 (yes) | λ clone | ND | Yes | Yes | Yes | IGLV1-44 | IGLJ3*02 | 38→P | ɣλ | 104 | 0.13 | 1082 | IGHV2-26 |
| p3 | Positive (λ) | 9 (yes) | λ clone | ND | Yes | Yes | ND | IGLV1-44 | IGLJ3*02 | 38→P | αλ | 35.4 | 0.38 | 7000 | |
| p5 | Positive (λ) | 3.5 (no) | λ clone | ND | No | No | Yes | IGLV1-44 | IGLJ3*02 | 38→P | αλ | 217 | 0.06 | 2725 | |
| p12 | Positive (PND) | 35 (yes) | ND | ND | No | No | Yes | IGLV1-44 | IGLJ3*02 | 38→S | αλ | 57.5 | 0.82 | 7800 | |
| p15 | Positive (λ) | 6 (yes) | λ clone | ND | Yes | Yes | Yes† | IGLV1-44 | IGLJ3*02 | 38→A | αλ | 72 | 0.21 | 9055 | |
| p16 | Positive (λ) | 5 (unknown) | λ clone | ND | No | Yes | Yes | IGLV1-44 | IGLJ3*02 | 38→A | ɣλ | 51.9 | 0.25 | 739 | |
| p23 | Negative | <5 (no) | No clone | No clone | Yes | No | Yes | IGLV1-40 | IGLJ3*02 | 40→N | No monoclonal Ig | 46 | 0.76 | 845 | |
| p24 | Negative | 4 (no) | ND | ND | Yes | Yes | Yes | IGLV1-40 | IGLJ3*02 | 40→N | — | 47 | 0.5 | 1300 | |
| p20 | Positive (k) | 2.3 (yes) | κ clone | ND | Yes | No | Yes | IGLV1-40 | IGLJ3*02 | 40→N | ɣλ and ɣk | 150 | 0.55 | 1400 | |
| p19 | Positive (PND) | 2 (yes) | λ clone | ND | No | Yes | Yes | IGLV1-40 | IGLJ3*02 | 40→N | αλ | 43.3 | 0.61 | 11 188 | |
| p21 | Positive (PND) | 22 (yes) | ND | ND | Yes | Yes | ND | IGLV1-40 | IGLJ3*02 or 01 or IGLJ2*01 | 40→Q | ɣλ | 134 | 0.09 | 1423 | |
| p22 | Positive (λ) | 3 (yes) | λ clone | ND | No | Yes | Yes | IGLV1-40 | IGLJ3*02 | 40→N | αλ | 32 | 0.74 | 7290 | IGHV2-70 |
| p25 | Positive (λ) | 0 (no) | λ clone | No clone | No | Yes | Yes | IGLV1-36 | IGLJ3*02 | — | ɣλ | 37.8 | 0.72 | 5000 | Other clone (IGLV2-14/IGLJ3*02) |
| p26 | Positive (λ) | 1 (no) | ND | λ | No | Yes | Yes | IGLV1-36 | IGLJ3*02 | — | No monoclonal Ig | 107 | 0.36 | 7550 | |
| p29 | Negative | 1% (no) | No clone | No clone | Yes | No | Yes | IGLV3-1 | IGLJ2*01 | — | ɣλ | 40.2 | 0.75 | 3275 | |
| p30 | Negative | <5 (no) | ND | No clone | No | No | Yes | IGLV3-25 | IGLJ1*01 | — | ɣλ | — | — | 1966 | |
| p27 | Positive (λ) | 2 (no) | λ clone | ND | Yes | No | Yes | IGLV3-1 | IGLJ3*01 | — | ɣλ | 35 | 0.45 | 1210 | |
| p31 | Negative | 1%-2.5% (no) | No clone | ND | Yes | No | No | No clone | No clone | — | No monoclonal Ig | 41 | 0.69 | 1700 | |
| p32 | Negative | 2 (no) | ND | ND | Yes | No | No | No clone | No clone | — | ɣλ | 51.3 | 0.46 | 1190 | |
| p33 | Negative | 3.5 (no) | No clone | ND | No | No | No‡ | No clone‡ | No clone‡ | — | ɣλ | 25.2 | 1.31 | 1146 | |
| p34 | Negative | 0.3 (no) | No clone | ND | Yes | No | No | No clone | No clone | — | ɣλ | 21.8 | 0.54 | 705 | |
| p35 | Negative | 2 (no) | No clone | ND | Yes | ND | No | No clone | No clone | — | αλ | — | — | 2550 | |
| p36 | Negative | 2 (no) | ND | ND | No | No | No | No clone | No clone | — | αλ | 27 | 1.33 | 665 | |
| Bone lesion sequencing | |||||||||||||||
| p7 | Negative | 2 (no) | ND | ND | Yes | Yes | Yes | IGLV1-44 | IGLJ3*02 | 38→A | ɣλ | 15.3 | 0.77 | 1419 | |
| p8 | Negative | 1.5 (no) | No clone | ND | Yes | Yes | Yes | IGLV1-44 | IGLJ3*02 | 38→A | ɣλ and αk | 55.6 | 0.99 | 3560 | |
| p18 | Positive (k) | ND | ND | κ | Yes | Yes | Yes | IGLV1-44 | IGLJ3*02 | 38→P | ɣk and ɣλ | 21.6 | — | 1100 |
ND, not done; PND, phenotyping not done.
Sequencing on both BM aspirates and bone lesions.
Clone found on lymph nodes.
In accordance with previous publications,5,6 IGLV1-40 or IGLV1-44 variable genes were overrepresented and identified in 25 of all patients (71%), and in 83% of patients in whom a λ-positive clone was detected by sequencing (25/30). An IGLV1-40 or IGLV1-44 monoclonal sequence was found in the 4 bone lesion biopsy samples. In 5 patients (14%), other IGLV genes were found, including 2 patients with a monoclonal IGLV1-36 variable gene, which is closely related to the IGLV1-44 germline gene (Table 1), and 3 patients with monoclonal IGVL3 variable genes that in our view were not related to the POEMS syndrome. Two of them presented bone lesions that are likely the source of the POEMS-related monoclonal plasma cells. Finally, no monoclonal λ LC was found in the BM of 6 patients (17%). Among them, none had a detectable PC clone using conventional cytologic and immunophenotypic analysis, but we finally detected, in patient 33 (atypical form of POEMS), an IGLV1-44/IGLJ3*02 clonal sequence in a paraffin-embedded lymph node biopsy, using Sanger sequencing on DNA (no RNA available from this sample). Ig RNA sequencing allowed the detection of an IGLV1-40 or IGLV1-44 monoclonal LC in the BM from 11/19 patients (58%) whose BM samples were previously negative with usual techniques or positive for a κ clone. Among these, 10 were analyzed by NGS (RACE-RepSeq), including 6 patients previously found negative with Sanger analysis. For 2 other patients with negative routine/conventional analysis, another λ LC clone was found. All the IGLV1-40, IGLV1-44, and IGLV1-36 variable genes were rearranged on the same IGLJ3*02 joining segment, as previously described.6 However, this rearrangement does not lead to a canonical CDR3, as CDR3 sequences were highly diverse (Figure 1). Interestingly, all the other clonal IGLVs found, not related to IGLV1-40, IGLV1-44, or IGLV1-36, were not associated with the IGLJ3*02 gene (Table 1). Finally, we sequenced IGHV genes in 5 patients and did not find any homogeneity or recurrent mutations (all 5 deriving from different IGHV germline genes; Table 1).
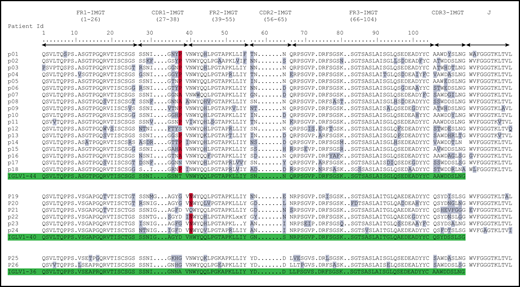
Deduced AA sequences of the monoclonal λ LC VJ domains in patients with POEMS syndrome compared with germline sequences according to IMGT numbering. Mutated AAs are highlighted in gray, and the redundant mutations in position 38 for IGLV1-44 and position 40 for IGLV1-40 are highlighted in red. Germline sequences are highlighted in green.
Deduced AA sequences of the monoclonal λ LC VJ domains in patients with POEMS syndrome compared with germline sequences according to IMGT numbering. Mutated AAs are highlighted in gray, and the redundant mutations in position 38 for IGLV1-44 and position 40 for IGLV1-40 are highlighted in red. Germline sequences are highlighted in green.
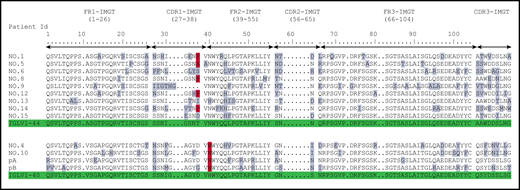
When compared with the corresponding germline sequences, the analysis of the IGLV1-40 or IGLV1-44 amino acid (AA) sequences revealed a novel specific mutational pattern with 2 redundant mutations at position 38 for IGLV1-44 and 40 for IGLV1-40 AA sequences, according to IMGT unique numbering27 (Figure 1). All (18/18; 100%) IGLV1-44 sequences are mutated in position 38, and the polar Threonine 38 was replaced by a hydrophobic residue in 16 of 18 sequences. A proline, which beside its hydrophobicity is known to disrupt protein secondary structures, was found in 12 (67%) of the 18 IGLV1-44 sequences, and an alanine in 4 other cases (22%), leading to a (P/A)VNWYQ consensus stretch in 61% of cases. Interestingly, a retrospective analysis of the series by Abe et al confirms the recurrence of this mutation, with 9 of 9 patients having a mutation in position 38, including 4 patients with a proline and 1 patient with an alanine6 (Figure 2). Because IGLV1-44 is frequently associated with AL amyloidosis, we analyzed sequences from previous studies by Perfetti et al,28 Comenzo et al,29 the Boston university AL-Base,30 and our own unpublished sequences, leading to a total of 100 IGLV1-44 sequences from patients with AL amyloidosis and 6 from other PC disorders. Strikingly, only 4 sequences presented a T→P/A mutation in position 38 (4%), and only 1 reconstituted the full (P/A)VNWYQ stretch (supplemental Figure 3). However, this V domain was not associated with the recurrent IGLJ3*02 domain. Regarding our IGLV1-40 AA sequences, the histidine 40 was replaced by an asparagine in 5 (83%) of 6 cases, and in the remaining case, the histidine was replaced by a glutamine. Interestingly, the asparagine residue in position 40 is present in the germline gene of the IGLV1-44 and IGLV1-36 domains and reveals a consensus VNWYQ stretch found in the FR2 of 73% of the IGLV1-36, IGLV1-40, and IGLV1-44 sequences (Figure 1). The H40→N substitution in the FR2 region in patients with IGLV1-40 clone is associated with a glutamine or aspartate in position 38 to form a (D/G)VNWYQ stretch that we also found in the 2 patients with IGLV1-36 clonal sequences. This mutation was retrospectively found in all monoclonal IGLV1-40 sequences (n = 4) from the series of Martin et al and Abe et al (Figure 2),5,6 but was observed in only 1 of the 18 IGLV1-40 sequences (5.6%) from AL amyloidosis and other PC disorders extracted from the AL-Base30 and our own database, and none reconstituted the full (D/G)VNWYQ consensus stretch.
Previously published AA sequences of POEMS syndrome compared with germline sequences according to IMGT numbering. Numbers 1 to 15 from Abe et al6 and pA/pB from Martin et al.5 Mutated AAs are highlighted in gray, and the redundant P or A mutations in position 38 for IGLV1-44 and N in position 40 for IGLV1-40 are highlighted in red. Germline sequences are highlighted in green.
Previously published AA sequences of POEMS syndrome compared with germline sequences according to IMGT numbering. Numbers 1 to 15 from Abe et al6 and pA/pB from Martin et al.5 Mutated AAs are highlighted in gray, and the redundant P or A mutations in position 38 for IGLV1-44 and N in position 40 for IGLV1-40 are highlighted in red. Germline sequences are highlighted in green.
Discussion
Despite the establishment of international criteria,1,3 the diagnosis of POEMS syndrome can be challenging because of the variety of clinical symptoms and frequent atypical clinical presentation lacking the full-blown spectrum of manifestations. Among these, even the main manifestation of the disease, polyneuropathy, is sometimes absent during early disease stage, and the level of VEGF, one of the main diagnostic criteria, may remain within normal range because of previous treatment by intravenous immunoglobulins or corticosteroids. In these situations, the diagnosis of POEMS is often delayed, resulting in delayed treatment introduction and increased morbidity. Indeed, the treatment of POEMS syndrome is highly specific and should be started before the apparition of a severe polyneuropathy. In the present study, we show that LC gene sequencing, particularly using the novel RNA-based NGS technique, is a potent tool to secure the diagnosis of POEMS, including patients with atypical presentation, and to guide therapeutic management.
In patients with plasmacytoma but negative BM using conventional analysis or in those lacking mandatory criteria for the diagnosis of POEMS, the search for a PC clone in the BM is crucial to deliver appropriate therapy. Based on the present series of POEMS sequences, we confirm the restriction of λ V domain usage to IGLV1-40 or IGLV1-44 rearranged to IGLJ3*02 gene, as already shown by others.5,6 We also found 2 patients with an IGLV1-36 variable gene; this gene could potentially be a new IGLV1 gene associated with POEMS syndrome, as it is known as a paralog of IGLVλ1-44 gene13,31 and contains the consensus sequence VNWYQ in the FR2 domain. Accordingly, an IGLV1-36 sequence associated with POEMS syndrome was recently found by RNAseq in the series of Nagao et al.32 Another hypothesis could be that the LC hypermutations that appear during B-cell response could be responsible for the assignment to an IGLV1-36-derived gene in place of the IGLV1-44 one. In any case, the association of this new λ V domain with POEMS syndrome remains to be confirmed with more sequencing data from patients with POEMS. Three other patients had unrelated IGLV clones (IGLV3-1 and IGLV3-25, none of them rearranged with the canonical IGLJ3*02 segment) that probably corresponded to incidental plasma cell clones not responsible for POEMS syndrome. Patients with POEMS syndrome frequently display hypergammaglobulinemia and increased polyclonal plasma cells in the BM and may have more than 1 monoclonal Ig.12 This was confirmed in our series, in which some patients had 2 concomitant IGLV clones and 2 were diagnosed with a κ LC clone in the BM (Table 1).
In 2018, Kawajiri-Manako et al showed for the first time that NGS may be useful to detect IGLV clones in BM from patients with POEMS.13 However, NGS-based Ig sequencing on DNA suffers from the frequent low PC burden observed in POEMS syndrome, and more generally in monoclonal gammopathies of clinical significance.2 Accordingly, significant clonal rearrangements were found in only 11 of 30 patients, and detection of some λ PC clones required heteroduplex analysis on cDNA.13 Our method proposes a sensitive assay based on cDNA analysis of Ig repertoire by NGS (RACE-RepSeq) allowing, in addition, the sequencing of the full-length VJ sequence. The sensitivity of the RACE-RepSeq approach is achieved thanks to the inherent high number of Ig transcripts in PC, accounting for up to 70% of total mRNA.33 As a consequence, even a very small fraction of monoclonal PCs, otherwise undetectable by conventional methods (including cytometry or DNA sequencing), is theoretically detectable using RACE-RepSeq.19 Accordingly, an IGLV clone was detected in 82% of BM analyzed, including from 13 patients in whom conventional methods did not allow the detection of an IGLV clone or detected an unrelated IGVK clone. This represents a 16% increase in the clone detection rate compared with that reported in a study of 87 patients from the Mayo Clinic.12 Obviously, the detection of an IGLV clone in the BM does not mean it is the causative LC, but the combination of restricted usage of IGLV genes, the unique IGLJ domain (IGLJ3*02) found in 100% of POEMS syndrome LC sequences, and the specific pattern of mutations revealed by full-length VJ sequencing may increase the diagnostic sensitivity. When we added the LC sequences of POEMS syndrome of the present work with the series of Abe et al6 and Martin et al,5 the H40→N substitution in the FR2 region of IGLV1-40 sequences was found in 9 (90%) of 10 patients, and the T38→P/A substitution in the CDR1 of IGLV1-44 sequences was found in 21 (78%) of 27 sequences. This appears highly specific to POEMS syndrome sequences, as previously highlighted in an abstract by Aravamudan et al,34 as such mutations were almost never observed in sequences from AL amyloidosis or other PC disorders (∼0.03% in a total of 124 IGLV1-40 and IGLV1-44 sequences), and none but 1 had a full PVNWYQ stretch found in IGLV1-44 sequences of POEMS syndrome, but not associated with the recurrent IGLJ3*02 domain. It would have been interesting to analyze retrospectively the clinical symptoms of this patient with unique AL amyloidosis with a clonal IGLV1-44 containing the PVNWYQ stretch (sequence DQ165740 in supplemental Table 1), as it could have been mistaken accidentally for an AL amyloidosis with neuropathy.35 Collectively, it seems that these recurrent mutations are highly specific to POEMS syndrome and may be a reliable diagnostic marker. As an example, patient 20 presented with a polyneuropathy, elevated VEGF, 158 mg/L λ free LC in the serum, and a plasmacytoma, but a κ+ monoclonal plasmocytosis in the BM and no λ LC clone found by Sanger sequencing in the BM. We considered that the BM plasmocytosis was not related to the POEMS syndrome, and the patient was treated with irradiation. The treatment was unsuccessful, and he subsequently received high-dose chemotherapy with autologous stem cell transplantation. Retrospectively, thanks to the RACE-RepSeq approach, we found in the BM an IGLV1-40 monoclonal sequence with the H40→N substitution associated with the IGLJ3*02 domain. The dissemination of this λ PC clone probably explains the ineffectiveness of irradiation. Conversely, patient 27 presented with an isolated plasmacytoma, a discrete elevation of λ-LC PCs in the BM, and a circulating monoclonal IgAλ. This patient was treated with irradiation and is still in clinical response after 8 years. We retrospectively performed RACE-RepSeq on the BM and detected an IGLV clone derived from the IGLV3-1 germline gene, which was not reported in POEMS syndrome, confirming the lack of association between the BM clone and the POEMS syndrome. We have also confirmed in this study that some patients with POEMS syndrome have more than 1 monoclonal Ig and that full-length VJ sequencing may help unveil the involved clone in such situation (p9).
Finally, this combination of restricted IGLV usage and a specific pattern of mutations can shed light on the mysterious pathophysiology of POEMS syndrome. The origin of the high level of VEGF, involved in many clinical features of the disease, is still controversial.3 Some studies have suggested that VEGF could be produced by monoclonal PCs in plasmacytoma36 or by both monoclonal and polyclonal PCs in BM.37 However, this hypothesis was refuted in a recent study showing that BM PCs of patients with POEMS do not produce more VEGF-A compared with monoclonal gammopathy of undetermined significance (MGUS) or multiple myeloma PCs.32 Given the low level of BM infiltration by monoclonal PCs in patients with POEMS, these controversies will likely be resolved by next-generation cytometry analysis or single-cell transcriptomic. One hypothesis is that the monoclonal λ LC could act as a paracrine or endocrine factor, which could activate the secretion of VEGF-A in cells/tissues. The restricted structure of monoclonal LCs from POEMS syndrome argues for an LC-Target interaction. Interestingly, when the PVNWYQ stretch was submitted to Blastp analysis,38 we found a 100% homology with the HSP90α chaperone protein known to be implicated in the regulation of angiogenesis through HIF1 chaperoning and to be a ligand for CD91 in its secreted form.39,40 Thus, we hypothesize that the λ monoclonal LC in POEMS syndrome could drive the secretion of VEGF by a mechanism of molecular mimicry with HSP90α.
In summary, using a new highly sensitive method of RNA repertoire sequencing (RACE-RepSeq), we confirmed the restricted usage of IGLV and IGLJ genes in POEMS syndrome, found a putative new germline gene (IGLV1-36), and highlighted peculiar patterns of mutations that could help to confirm the diagnosis and will likely shed new lights in the pathophysiology of this complex disease. Although it deserves to be confirmed with largest cohorts and with adequate controls, the sensitivity of RACE-RepSeq seems to outcompete other methods to detect small PC clones. The application and the benefits of RACE-RepSeq in other monoclonal gammopathies of clinical significance, or in the detection of minimal residual disease in PC disorders in general, deserve to be investigated.
Data are available at ENA European Nucleotide Archive database under accession number PRJEB35157.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors acknowledge the use of the Boston University ALBase, supported by National Institutes of Health, National Heart, Lung, and Blood Institute grant HL68705, in this work.
This work is supported by Ligue Nationale Contre le Cancer du Limousin and Fondation Française pour la Recherche contre le Myélome et les Gammapathies monoclonales. S.B., D.L., and F.A. are supported by the French Ministry of Research “Plan maladies rares.” M.V.A. is supported by Fondation ARC pour la Recherche sur le Cancer.
Authorship
Contribution: S.B. performed and analyzed experiments and wrote the manuscript; V.J., M.F., V.P., M.C., and F.B. critically reviewed the manuscript; A.S. and M.A. collected and analyzed data and reviewed the manuscript; N.G. collected and analyzed data; M.V.A. drafted the manuscript; D.L. and F.A. collected clinical data; and C.S. and A.J. supervised this study, analyzed experiments and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Christophe Sirac, UMR CNRS 7276/INSERM U1262, CBRS-Faculté de Médecine, 2 rue du Dr Marcland, 87025 Limoges, France; e-mail: christophe.sirac@unilim.fr; Arnaud Jaccard; CHU de Limoges, Service d’hématologie clinique, 2 rue Martin Luther King, 87000 Limoges, France; e-mail: arnaud.jaccard@chu-limoges.fr.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal