Background: Recent promising results from the CD33-directed immunoconjugate, gemtuzumab ozogamicin (GO), has transformed the realm of acute myeloid leukemia (AML) treatment strategies and has opened the door for several other CD33-directed immunotherapeutic strategies to be tested in clinical settings. Starting from accelerated approval in 2000, GO was discontinued in 2010 due to high levels of death in a major phase 3 clinical trial until follow-up efficacy studies allowed for its re-approval in 2017. Despite the complex path of GO as a therapeutic, its impact in the field has been unprecedented and thus ongoing studies seek to improve treatment strategies using GO. Among the most prevalent challenges to this goal is the variation in structure and expression levels of CD33. In particular, previous studies from our group reported rs12459419 C>T, a splicing SNP, resulting in formation of a shorter CD33 isoform (CD33D2) that lacks the critical immunogenic IgV domain (Lamba, 2017). Given that GO recognizes the IgV domain, its therapeutic effect is compromised in patients expressing CD33D2 isoform (CT and TT genotypes). The lack of benefit observed in rs12459419 heterozygous patients is perplexing and warrants in depth investigations to understand biology of CD33D2 isoform. However, the lack of a specific antibody that can recognize the CD33D2 isoform is a significant roadblock to advancing the: (1) understanding of CD33D2 biology; (2) evaluation of CD33D2 isoform as a novel drug target; and (3) explanation regarding why heterozygous patients have compromised response to GO, despite having intermediate levels of CD33FL isoform.
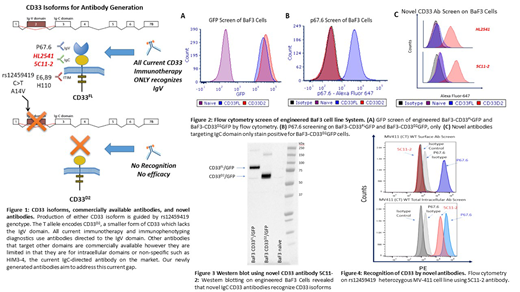
Methods: To overcome this roadblock, we focused on developing novel CD33 antibodies specifically targeted to the shared IgC domain between both isoforms (Figure 1). In brief, peptides spanning IgC domain of CD33 were used as immunogens in either Balb/c or C57BL/6 mice for antibody generation. The novel antibodies, named 5C11-2 and HL2541, were used to assess surface-level, intracellular expression, and intracellular localization of CD33 isoforms. The newly developed IgC targeting antibodies and the well-established IgV targeting P67.6 antibody were tested by flow cytometry and western blotting on the CD33 negative BaF3 cell line and BaF3 cells engineered to express CD33FL (BaF3-CD33FLGFP) and CD33D2 (BaF3-CD33D2GFP), as well as AML cell lines representing different rs1245419 genotypes.
Results: As expected, naïve Baf3 cells showed no GFP fluorescence signal whereas BaF3-CD33FLGFP, BaF3-CD33D2GFP were positive for GFP confirming expression of CD33 isoforms (Figure 2A). Subsequent staining with P67.6 only produced a positive signal in BaF3-CD33FLGFP cells while BaF3-CD33D2GFP, which lack the IgV domain, were negative (Figure 2B). Interestingly both novel IgC targeting antibodies, 5C11-2 and HL2541, only stained positive for BaF3-CD33D2GFP cells and were negative for BaF3-CD33FL cells (Figure 2C). Western blotting confirmed the recognition of both CD33FL and CD33D2 isoforms by the IgC targeting novel antibody 5C11-2 (Figure 3). These results indicate a potential steric hindrance caused the bulky nature of the clustered extracellular Ig-folds that may have impacted recognition of native CD33FL by IgC targeting antibodies. Cytometry analysis using 5C11-2 on MV4-11 cells, which are heterozygous for the rs12459419 SNP, showed fluorescence signal when stained IgV specific P67.6 but were negative for the IgC specific novel antibody (Figure 4). These results warrant further investigation to establish the exact localization of CD33D2 in context of different cell types and primary AML cells from patients. Nonetheless, our results report successful development of novel CD33 antibodies that recognizes CD33D2 isoform, which holds promise in establishing CD33 biology and understanding the therapeutic implications such as mechanisms contributing to compromised response in rs12459419 heterozygous patients.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal