Introduction: The myelodysplastic syndromes (MDS) are a group of clonal haematopoietic stem cell disorders and they can be a consequence of genomic/chromosomal instability. The World Health Organization (WHO) 2008 classification divides the MDS in 4 types: refractory anaemia with excess of blast (RAEB), refractory anaemia (RA), refractory cytopenia with multilineage dysplasia (RCMD) and chronic myelomonocytic leukaemia (CMML). Various mechanisms contribute to the pathogenesis and prognosis of the disease and currently next generation sequencing (NGS) detects pathogenic gene mutations that can allocate patients to different risk classes. MicroRNAs (miRNAs) are small non-coding RNA molecules that regulate gene expression via post-transcriptional mechanisms and may have oncogenic properties or act as tumour suppressor and have an active role in the onset of myeloid disorders. A prospective study based on integration of NGS and miRNA quantification was launched at Policlinico San Martino in 2017. The aim of this paper is to report the results and the correlation with the clinical features.
Patients and methods: 28 consecutive patients were enrolled in this study because they met our eligibility criteria: (i) availability of bone marrow sample at diagnosis, (ii) IPSS and (iii) clinical follow up. Specifically, 11 patients were affected by RAEB, 9 by RA, 5 by RCMD and 3 by CMML. Median age was 66 years old (48 - 85). Eight patients evolved into acute myeloid leukaemia. NGS analysis was performed with Myeloid Oncomine Panel Thermo fisher; miRNA expression was quantified with TaqMan Advanced miRNA Cards. In the context of a machine learning supervised analysis we evaluated if there were significant differences in the miRNAs expression among the patients classified based on the WHO 2008 classification. This analysis was conducted using l1l2-penalized regularization method within PALLADIO, a machine-learning framework that can provide robust variable selection in high-dimensional problems. The identified miRNA signature was characterized though a miRNA enrichment analysis using the webtoolkit WebGestalt.
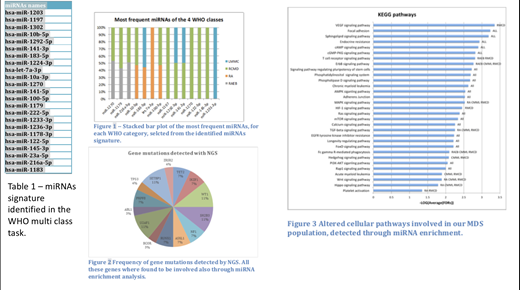
Results: each patient carried a distinctive miRNA signature composed by a median of 650 miRNA. The supervised analysis identified a signature of 24 miRNAs (table 1) able to discriminate the 4 WHO classes with a balanced accuracy of 0.616 (whose significance with respect to the balanced accuracy distribution of the permutation batch experiments has 2.206e-08 p-value). Among these miRNAs, 14 were selected because frequently up-regulated in most the patients of one or more WHO classes (figure 1). NGS identified mutations in 43% of patients affected by RAEB, RCMD and CMML in genes known to be pathogenic for MDS (figure 2). miRNA enrichment analysis allowed the identifications of genes involved in (i) pathways (some in common and some specific for a WHO class) likely involved in the onset of MDS, (ii) in diseases related to MDS and to disease evolution into acute myeloid leukaemia. Some of these genes were the same with the ones identified by NGS in our study population (i.e. TET2, ASLX1, RUNX1, IKZF1, SRSF2) but many others (e.g., GATA2, NfkB, BCL2, CD46, mTOR) were not detected by NGS but are likely to be involved in MDS pathogenesis and disease progression as described previously by other groups (figure 3).
Conclusions: our study gives a further input toward the understanding of pathogenesis in MDS through the integration of miRNA analysis with NGS at the diagnosis of MDS. With the identification of specific miRNA signature for each patient it is possible to highlight multiple genes pathway that may contribute to the onset and progression of MDS. In a prospective trial we also aim to verify if a specific miRNA signature is linked to azacitidine resistance.
Mufti:Celgene: Consultancy, Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal