Background: Acute myeloid leukemia (AML) with double-mutated CEBPA (dmCEBPA) occurred in about 10% of AML and it is usually associated with normal karyotype AML. It is classified as a favorable risk AML with an estimated 5 years overall survival in excess of 50%. Patients who achieved complete remission (CR) with induction chemotherapy will be consolidated with chemotherapy. Around 40% of the patients would suffer a relapse and required further salvage chemotherapy followed by allogeneic stem cell transplant. Co-mutations occurred frequently in dmCEBPA AML with TET2 being the most commonly co-mutated (34%) followed by GATA1 mutation (21%), WT1(13.7%), DNMT3a (9.6%), ASXL1 (9.5%), NRAS (8.4%) and other less common genes. Only TET2 mutation appeared to have negative impact on prognosis. In this study, we explore the relapse cases with Whole-Exome sequencing (WES) and compared with a control case (Chemo-responsive dmCEBPA AML who is in continuous complete remission (CCR)). We aimed to identify novel mutations not previously reported and with potential as new prognostic marker as well as therapeutic target for further exploration.
Method: We identified 3 relapsed dmCEBPA AML cases from our leukemia cell bank and compared with a case of chemo-responsive dmCEBPA AML who is still in CCR at 8 years follow-up. Duration of prior CR for case #1, #2 and #3 were 10 months, 1 year 9 month and 1 year 10 months. WES was performed for all 4 cases. In each case, the variants were annotated using the IonReporter software and an additional customized script that annotates for variant population frequency, genotype-phenotype associations and computational missense prediction tools. Variants were filtered by removing all reference calls, intergenic variants and variants not present in the chemoresponsive case. Only exonic and non-synonymous variants were considered. Next, we look for variants with a phenotype-genotype association common with the AML phenotype by looking for variants with "Leukemia" terms listed in OMIM. In addition, in order to identify novel mutations not presently associated with AML, we identified variants that had a "Pathogenic" entry in ClinVar.
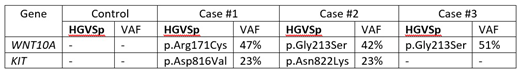
Results: All 4 cases revealed the typical CEBPA mutants with VAF exceeding 40% except one with germline mutation. When compared with the chemo-responsive case, KIT was mutated in 2 out of 3 relapse cases. WNT10A was mutated in all 3 cases. The details of the mutated genes are presented in Table 1 (see below).
WNT10A gene encodes for a secretory signaling protein in the WNT family of structurally related genes that are involved WNT-beta-catenin-TCF signaling pathway. WNT10A is strongly expressed in cell lines of promyelocytic leukemia. Beta-catenin, the downstream target of the WNT signaling pathway, is highly expressed in poor prognostic AML cells (Staal et al. Nature review Immun 2008). The Gly213 residue is highly evolutionary conserved in vertebrate with a PhyloP score of 9.8. Both missense variants Gly213Ser and Arg171Cys are computationally predicted be damaging (DANN score 0.99). However, these 2 variants are fairly common in the East Asian population (gnomAD_EAS MAF G213S 2.7% & R171C 1.5%). Both variants are ACMG guidelines classified as Variants of Unknown Significance (VUS).
KIT gene encodes for the proto-oncogene c-kit. Mutations in c-kit are associated with mastocytosis and AML. Both variants occur within the kinase domain (exon 17) of KIT. Asp816Val variant has been linked to poorer prognosis and worse outcome on AML patients. Both missense variants are computationally predicted to be damaging (DANN score 0.99). Both Asp816Val and Asn822Lys are AMCG classified as Likely Pathogenic and VUS respectively.
Conclusion: With stringent criteria of filtering in WES of relapsed dmCEBPA cases, when compared with the control, we found that KIT was mutated in 2 out of 3 cases, while WNT10A was mutated in all 3 cases. Though the sample number was small, these findings would warrant further evaluation in larger cohort of dmCEBPA AML and interrogation in pre-clinical model.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal