BACKGROUND :
Clinical Phenotype and outcomes of patients with Acute myeloid leukemia (AML) in the Indian subcontinent differs from published literature. A younger age at diagnosis and higher induction mortality complicate AML management in India(1). Metaphase Karyotyping represents the backbone of prognostication and risk stratification in AML. Optimal treatment strategies for the cohort of Cytogenetically normal AML are still under evaluation. Applications of Next generation Sequencing (NGS) techniques in AML have unravelled the genetic heterogeneity of this disease. Whole genome sequencing has identified many novel mutations leading to tremendous improvements in diagnosis and risk stratification. Development of therapies targeting these genetic alterations is enabling a gradual shift from non-specific approaches to personalised therapy tailored to an individual patient's genome. This will undoubtedly translate to better clinical outcomes for this disease, with otherwise poor prognosis. Whole genome sequencing is still in a nascent stage in Indian settings with no published literature on genomics in AML till date. We aimed to study the genomic landscape of AML in the Indian population and to co-relate this with clinical outcomes over the course of 1 year.
METHODS:
We recruited 34 newly diagnosed patients with AML who presented to our Centre (Mazumdar Shaw Medical Centre, Narayana Health City, Bangalore, India) between November 2017 and May 2018. Clinical and laboratory details of all patients were recorded. Bone marrow and paired peripheral blood samples were drawn before initiating therapy. Whole genome sequencing and Exome capture was done for each sample using Ilumina HiSeq platform. Patients were risk stratified as per ELN 2017 and treated as per NCCN guidelines. Patients were followed up prospectively for one year from initial diagnosis. Genetic results were stratified according to gene function and analysed with respect to predefined clinical outcomes (remission status post induction, relapse rates, progression free and overall survival).
RESULTS:
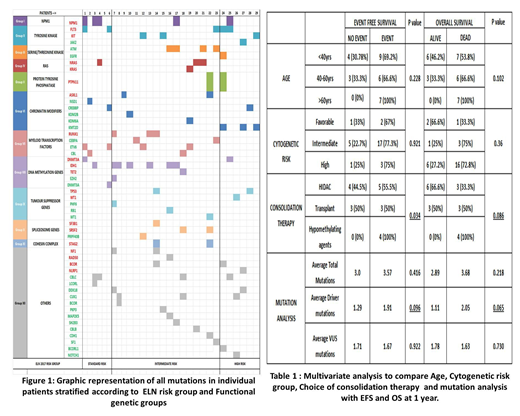
Amongst the 34 study participants, 5 patients failed QC during sequencing and were de-recruited. Hence 29 patients were available for final analysis. Median age of patients was 42 years with 13 patients (44.8%) less than 40 years of age.18 patients (60%) had normal cytogenetics at baseline.17 patients (58%) were classified as intermediate risk and 6 patients each as Standard and high risk, as per ELN 2017. 22 patients (79.3%) patients received standard Induction chemotherapy (3+7 regimen) while 6 patients received hypomethylating agents. Overall CR rate following induction at Day 28 was 50% and Induction mortality was 21.42%. 6 patients underwent an Allogenic Stem cell transplant.
A total of 96 mutations (47 driver and 49 VUS mutations) in 123 genes were identified. The average number of Driver mutations was 1.48 per patient. IDH genes were the most frequently mutated Driver genes followed by FLT3 mutations. Frequency of NPM1 mutations was significantly low (17.25%). Highest frequency of VUS mutations was seen in the ETV6, ATM and CBLC genes. Highest frequency of somatic mutations were identified in the genes encoding for myeloid transcription factors and DNA methylation. Average driver mutations showed significant co-relation to Age (> 60 years) and high burden of Bone marrow blasts (>30%). An updated risk stratification incorporating mutation analysis findings resulted in re-stratification of 8 intermediate risk patients into high risk. 2 patients with detectable FLT3 ITD mutation by NGS were negative by PCR. Choice of consolidation therapy and Driver mutation status were found to show statistically significant association with both Event free survival and Overall survival at 1 year. Increased driver mutation burden was associated with increased refractoriness to chemotherapy and poor EFS and OS. Mutations in Tumour suppressor genes, were associated with suboptimal treatment outcomes and poor survival.
CONCLUSIONS
Genomic landscape of AML in Indian patients shows significant differences from published literature. This may hold clues to the differing biological characteristics of AML seen in this population. Genome based risk stratification and tailored therapy needs to be adapted into the management of AML. This data provides valuable insights into developing therapeutic strategies for Indian patients.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal