Background:
More than 45 mutations have been identified in association with HR-MDS. In the majority of patients with MDS (80%) co-mutations are present and the prognostic contribution of each individual mutation remains elusive, especially after adjusting for clinical variables such as IPSS-R score. N-RAS and K-RAS mutations as well as regulators of the Ras pathway (e.g. PTPN11, NF1) are frequently observed (15-20%) in HR-MDS, however their clinical impact is unclear, especially in de novo MDS. Rigosertib (RGS) is a non-ATP-competitive small molecule RAS mimetic that has the potential to block RAS-RAF-MEK-ERK and PI3K-AKT-mTOR signaling pathways (Athuluri-Divakar 2016). Rigosertib has the potential to also inhibit wildtype upregulation of RAS. We report here genomic profiling at the time of study entry in the ongoing phase 3 randomized global study (known as INSPIRE) in patients with HR-MDS after failure of HMA therapy.
Methods:
INSPIRE (NCT02562443) is a global randomized Ph3 trial in pts with HR-MDS after HMA failure with an overall target enrollment of 360 pts with currently 298 pts randomized. Pts are randomized 2:1 to rigosertib or physician's choice of treatment. The primary endpoint is overall survival (OS). All pts failed to respond or progressed on HMA therapy. Key inclusion criteria includes: age < 82 years, RAEB-1, RAEB-2 or RAEB-t; intermediate risk (IR), high risk (HR) and very high risk (VHR) per IPSS-R; ≥ 1 cytopenia; duration of prior HMA ≤ 9 cycles within 12 months; last dose of HMA ≤ 6 months before enrollment; baseline blast counts between 5-29% and one of the following: progression any time after initiation of HMA treatment, intolerance to HMA, failure to achieve complete remission (CR), partial remission (PR), or hematologic improvement (HI) after six 4-week cycles of AZA or either four 4-week or four 6-week cycles of DAC, or relapse after initial CR, PR or HI. Bone marrow samples were collected at study baseline and throughout the study for mutational analysis as an exploratory endpoint. Baseline blast counts are described as % reported in bone marrow aspirate at screening. Genomic DNA was extracted from diagnostic bone marrow or peripheral blood samples and targeted capture deep sequencing of 295 genes were performed (median sequencing depth 500x) using Agilent's SureSelect custom panel. Modified Mutect and Pindel were used to identify high-confidence somatic mutations.
Results:
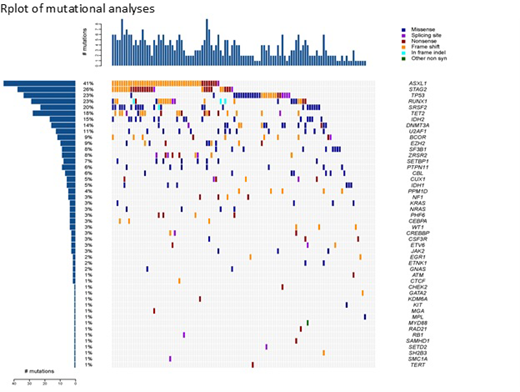
All data is presented as blinded aggregate results for both arms of the study. Baseline mutations are presented for 114 pts; 92 pts were randomized and 22 pts were screen failures. Median age is 72 years (59-81). The IPSS-R scores for the pts randomized were: Intermediate 5(5%), High 30 (32%) and VHR 56 (60%). In total 48 different mutations were identified at baseline prior to pts receiving study treatment. The most common mutations identified in pts were ASXL1 40%, STAG2 26%, TP53 24%, RUNX1 22%, SRSF2 and TET2 each 18%. Average number of mutations per pt was 3.06. At baseline, 4 pts (4%) had no mutations, 17 pts (18%) had only 1 mutation while 17 pts (18%) had between 6-8 mutations. N-RAS and K-RAS mutations occurred in 4 pts each (7% of pts) and all in the presence of other mutations. Mutations involving regulators of the Ras pathway (N-RAS, K-RAS, PTPN11, NF1) occurred in 12 (13%) of patients. Of note, 23 pts (25%) had IDH1 (6 pts) or IDH2 (17 pts) and the majority of these pts (68%) were High/VHR with an average blast count of 15% (range 5-27%) at study entry. The high proportion of IDH1/2 mutations observed is most likely due to inclusion of patients with RAEB-t. These results will be updated at the meeting with blinded baseline mutational analyses for all pts randomized into the study.
Conclusion:
The baseline mutational analyses from the INSPIRE study provides important initial insights into the genomic profile of pts with HMA failure, especially for the subset with VHR. Following analysis of the primary endpoint, it is anticipated that correlation of overall survival and clinical response with mutational status will be possible, including changes in mutations following therapy. Given the number of mutations involving the Ras pathway the efficacy of rigosertib in patients with this group of mutations will also be examined.
Garcia-Manero:Amphivena: Consultancy, Research Funding; Helsinn: Research Funding; Novartis: Research Funding; AbbVie: Research Funding; Celgene: Consultancy, Research Funding; Astex: Consultancy, Research Funding; Onconova: Research Funding; H3 Biomedicine: Research Funding; Merck: Research Funding. Luger:Agios: Honoraria; Jazz: Honoraria; Kura: Research Funding; Onconova: Research Funding; Pfizer: Honoraria; Seattle Genetics: Research Funding; Daichi Sankyo: Honoraria; Genetech: Research Funding; Cyslacel: Research Funding; Ariad: Research Funding; Biosight: Research Funding; Celgene: Research Funding. Al-Kali:Astex Pharmaceuticals, Inc.: Research Funding. Jedrzejczak:Amgen: Consultancy, Other: travel support for hematology meetings (ASH, EBMT, EHA) ; Celgene: Other: travel support for hematology meetings (ASH, EBMT, EHA) ; Takeda: Consultancy; Novartis: Research Funding; Roche: Other: travel support for hematology meetings (ASH, EBMT, EHA) . Díez-Campelo:Novartis: Consultancy, Membership on an entity's Board of Directors or advisory committees, Research Funding; Celgene Corporation: Consultancy, Membership on an entity's Board of Directors or advisory committees, Research Funding. Zbyszewski:Onconova Therapeutics, Inc.: Employment. Cavanaugh:Onconova Therapeutics, Inc.: Employment. Woodman:Onconova Therapeutics, Inc.: Employment. Fruchtman:Onconova Therapeutics, Inc.: Employment. Takahashi:Symbio Pharmaceuticals: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal