Introduction
Waldenstrom macroglobulinemia (WM) is a rare, incurable, indolent non-Hodgkin lymphoma that is preceded by an IgM monoclonal gammopathy of undetermined significance (IgM MGUS). In recent years, better understanding of the pathobiology of WM and IgM MGUS has strengthened the hypothesis that they may represent different stages of the same disease. Studying the relationship between these two conditions is the essential first step to understanding the mechanisms driving the malignant transformation from IgM MGUS to WM. Recently long non-coding RNAs (lncRNAs) have garnered significant recognition for their roles in the regulation of gene expression and cellular function. lncRNA can regulate gene expression of neighboring protein-coding genes through multiple mechanisms such as by recruiting chromatin modifiers and increasing accessibility of genes to transcription proteins, or by directly binding to enhancers or promoters. Here we present the first analysis of the lncRNA expression profile in patients with WM compared to IgM MGUS.
Methods
Twenty-one patients with untreated WM (n=14) and IgM MGUS (n=7) seen at Mayo Clinic were included in this analysis. Malignant cells were isolated from bone marrow samples using CD19 and CD38 positive selection. Total RNA was extracted and sequencing was performed using the Illumina HiSeq4000 NGS platform. Differentially expressed lncRNAs in WM compared to IgM MGUS were defined as log2 fold change (FC) > 1.0 (upregulated) or < -1.0 (downregulated) and a false discovery rate (FDR) < 0.05. The nearest gene to each lncRNA was also identified and their gene expression was detected. Using the Ingenuity Pathway Analysis software and taking into account the transcriptional expression and known function of these genes, a correlation between cell function and disease analysis was performed.
Results
A total of 119 lncRNAs were differently expressed between WM and IgM MGUS (17 transcripts were upregulated in WM, while 102 were downregulated in WM). Sixteen unique genes were identified as the nearest gene to 17 upregulated lncRNAs, while 18 genes were identified as the nearest genes to the top 18 downregulated lncRNAs in WM compared to IgM MGUS. Several of the upregulated lncRNA in WM were in proximity to known oncogenes.
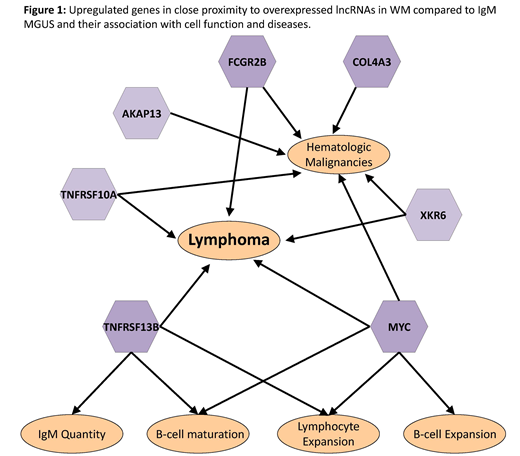
The well-known oncogenic lncRNA PVT1 was found to be upregulated in WM compared to IgM MGUS (log2 FC 1.35, FRD 0.04). PVT1 is overexpressed in several malignancies and associated with inhibition of apoptosis and increased cell proliferation. PVT1 is in close proximity to MYC and co-expression of PVT1-MYC has been described in multiple cancers. PVT1 also potentiate MYC amplification by increasing MYC protein stability, therefore promoting malignant cell growth.
The lncRNA TCB1D27, also upregulated in WM compared to IgM MGUS, is located in close proximity to the oncogene TNFRSF13B (TACI), which was found to be overexpressed in WM patients (log2 FC 1.61, FDR <0.001). TACI is a receptor for BLyS/BAFF, which modulates the development of normal B-cells and plays a role in various B-cell malignancies. In fact, BLyS/BAFF level is known to be increased in WM and to promote survival and proliferation of WM B cells.
The upregulated lncRNA RP11-25K21.1 in WM is located in close proximity to the oncogene FCGR2B (CD32B), which was also upregulated in WM compared to IgM MGUS (log2 FC 1.59, FDR 0.001). Overexpression of CD32B was previously shown to be associated with inferior outcomes in DLBCL and FL patients, presumably secondary to an increased internalization and clearance of rituximab due to increased CD32B expression on malignant cells.
Several other upregulated lncRNAs in WM compared to IgM MGUS are also in proximity of other oncogenes such as PCED1B, POLD3, CLLU1, and SMAD7.
Finally, we considered the expression of the nearest genes and their reported cell function and association with diseases. Several genes that are known to have a role in the cell proliferation and/or neoplasm development were noted to be upregulated in WM (figure 1).
Conclusion
We present the first report of lncRNAs expression in WM and IgM MGUS in addition to a concurrent analysis of the gene expression of the nearest protein-coding genes. Several known oncogenes are in proximity of overexpressed lncRNAs in WM compared to IgM MGUS that could indicate potential implications in the pathobiology of these diseases and the mechanism driving the malignant transformation to WM.
Paludo:Celgene: Research Funding; Celgene: Research Funding; Verily Life Sciences: Research Funding; Verily Life Sciences: Research Funding. Dasari:The Binding Site: Patents & Royalties: US Patent Rights on Mass Spectroscopy Licensing agreement with The Binding Site, Research Funding. Kapoor:Celgene: Honoraria; Cellectar: Consultancy; Janssen: Research Funding; Sanofi: Consultancy, Research Funding; Amgen: Research Funding; Takeda: Honoraria, Research Funding; Glaxo Smith Kline: Research Funding. Ailawadhi:Amgen: Consultancy, Research Funding; Pharmacyclics: Research Funding; Takeda: Consultancy; Janssen: Consultancy, Research Funding; Cellectar: Research Funding; Celgene: Consultancy. Gertz:Amyloidosis Foundation: Research Funding; i3Health: Other: Development of educational programs and materials; Medscape: Consultancy, Speakers Bureau; Ionis/Akcea: Consultancy; Celgene: Consultancy; Appellis: Consultancy; Amgen: Consultancy; Teva: Speakers Bureau; Alnylam: Consultancy; Prothena Biosciences Inc: Consultancy; Janssen: Consultancy; Spectrum: Consultancy, Research Funding; Annexon: Consultancy; Physicians Education Resource: Consultancy; Abbvie: Other: personal fees for Data Safety Monitoring board; Research to Practice: Consultancy; Johnson and Johnson: Speakers Bureau; DAVA oncology: Speakers Bureau; Pharmacyclics: Membership on an entity's Board of Directors or advisory committees; Proclara: Membership on an entity's Board of Directors or advisory committees; Springer Publishing: Patents & Royalties; International Waldenstrom Foundation: Research Funding. Novak:Celgene Coorperation: Research Funding. Ansell:Affimed: Research Funding; Regeneron: Research Funding; Mayo Clinic Rochester: Employment; Seattle Genetics: Research Funding; Bristol-Myers Squibb: Research Funding; Bristol-Myers Squibb: Research Funding; Affimed: Research Funding; LAM Therapeutics: Research Funding; Bristol-Myers Squibb: Research Funding; Seattle Genetics: Research Funding; Affimed: Research Funding; Affimed: Research Funding; Mayo Clinic Rochester: Employment; Bristol-Myers Squibb: Research Funding; Mayo Clinic Rochester: Employment; Seattle Genetics: Research Funding; Trillium: Research Funding; Mayo Clinic Rochester: Employment; Bristol-Myers Squibb: Research Funding; Bristol-Myers Squibb: Research Funding; Trillium: Research Funding; Regeneron: Research Funding; Seattle Genetics: Research Funding; Mayo Clinic Rochester: Employment; Trillium: Research Funding; Trillium: Research Funding; Mayo Clinic Rochester: Employment; Regeneron: Research Funding; Regeneron: Research Funding; LAM Therapeutics: Research Funding; Mayo Clinic Rochester: Employment; Seattle Genetics: Research Funding; LAM Therapeutics: Research Funding; Seattle Genetics: Research Funding; Mayo Clinic Rochester: Employment; LAM Therapeutics: Research Funding; LAM Therapeutics: Research Funding; Trillium: Research Funding; Affimed: Research Funding; Trillium: Research Funding; Regeneron: Research Funding; Affimed: Research Funding; Affimed: Research Funding; Bristol-Myers Squibb: Research Funding; LAM Therapeutics: Research Funding; Trillium: Research Funding; LAM Therapeutics: Research Funding; Regeneron: Research Funding; LAM Therapeutics: Research Funding; Trillium: Research Funding; Mayo Clinic Rochester: Employment; Regeneron: Research Funding; Bristol-Myers Squibb: Research Funding; Affimed: Research Funding; Seattle Genetics: Research Funding; Affimed: Research Funding; Regeneron: Research Funding; Seattle Genetics: Research Funding; Bristol-Myers Squibb: Research Funding; LAM Therapeutics: Research Funding; Regeneron: Research Funding; Seattle Genetics: Research Funding; Trillium: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal