Introduction: Current classifications of low-grade B-cell lymphomas (LGL), including splenic marginal zone lymphomas (SMZL), nodal marginal zone lymphomas (NMZL), and Lymphoplasmacytic lymphomas (LPL) are based on a mixture of clinical features and morphologic, immunophenotypic, and genetic findings from the tumor biopsy specimen. While this approach to classification makes pathologic diagnosis more precise, the corresponding clinical impact for the timing and choice of treatment is limited, and differentiating between cases can be challenging. Although LGL are considered indolent and the 10-year overall survival (OS) is about 80%, 70% of cases will eventually require treatment and approximately 30% of patients display a more aggressive phenotype and have a poor prognosis. Consequently, further investigations into the driving genetic, biological, and immune mechanisms of LGL are essential for early identification of high-risk patients and design of personalized treatments.
Materials and Methods: RNA-seq was performed on 63 newly diagnosed LGL patient samples from the Mayo Clinic/University of Iowa Lymphoma SPORE: SMZL (N=48), NMZL (N=6), LPL (N=5), MALT (N=2), and BCL (N=2) as well as 5 normal memory B cell controls (CD19+CD27+). For identification of biologic clusters, filtered RNA-seq data was analyzed using the Non-negative Matrix Factorization (NMF) clustering tool from the Broad Institute against the normal samples. Each cluster was analyzed for differential gene expression. This analysis generated cluster-specific T values for each gene. Genes that were significantly associated with a cluster (FDR<0.05) were analyzed for genetic ontology. Cibersort was used to deconvolute the tumor microenvironment (TME). We performed whole exome sequencing (WES) analyses on 60/63 matched LGL to characterize mutations and copy number alterations.
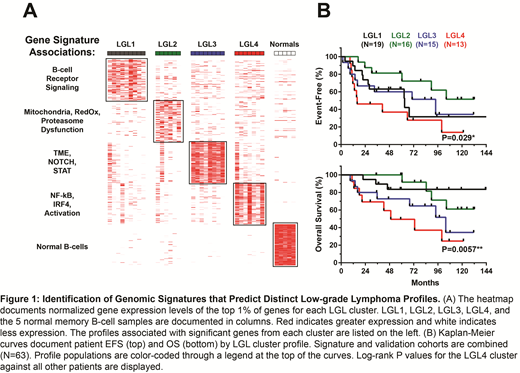
Results: WES revealed the presence of previously reported mutations in genes such as MYD88, SPEN, NOTCH, and KLF2 as well as copy number alterations 7q31.2, BCL6, TNFAIP3, and BCL2. NMF clustering of RNA-seq data resulted in a best fit of 5 clusters, named LGL1 through LGL4 and "Normals". Pathologic subtypes were not exclusive to a specific cluster. Differential RNA expression analysis resulted in gene sets that were differentially expressed in each cluster. A cluster signature of the top 1% of associated genes (N=94) was created for each. LGL1 genes were associated with higher BCR signaling. LGL2 was characterized by regulatory dysfunction, particularly concerning mitochondrial integrity. LGL3 genes were associated with high TME and NOTCH and STAT signaling components. Specifically, LGL3 was associated with high CD8+ T-cell and M2 macrophage TME presence. LGL4 was distinguished by the presence of aggressive genetic programs related to B-cell activation such as NF-kB and IRF4. Consequently, LGL4 patients had significantly poorer rates of event-free survival (EFS) (P=0.029) and OS (P=0.006) when compared against the other groups. The top 1% gene signature (N=94) was next validated against an independent cohort of 84 LGL samples with gene expression data (SMZL [N=34], NZML [N=24], and LPL [N=24]). A similar cluster pattern emerged, with pathological subtypes distributing across LGL classifications based on respective transcriptomic signatures. Association of DNA variants within each cluster was also analyzed, but due to the low frequency of genomic variants detected in LGL, nothing was significantly associated. However, we did see an enrichment of BCL2, BCL6, and TNFAIP3 alterations in LGL4 and an overall lower driver gene mutation frequency in LGL2.
Conclusions: Gaining a greater understanding of LGL based on their genetic, biologic, and immune profiles will establish a valuable platform for clinicians to identify high-risk cases and make better therapeutic decisions. Using a large cohort of well-annotated cases we identify novel clusters of LGL that present unique genomic and clinical profiles. Our study lays the groundwork for a precision therapy approach in LGL in which DNA or RNA profiles can be used to identify patients early in treatment who may not benefit from the current standard of care and who would benefit from closer monitoring and targeted agents.
Anagnostou:American Society of Hematology, Mayo Clinic/Iowa Lymphoma SPORE, Mayo Clinic Immune Monitoring Core, Mayo Clinic Hematology Small Grant: Research Funding. Ansell:Bristol-Myers Squibb: Research Funding; Regeneron: Research Funding; Regeneron: Research Funding; Seattle Genetics: Research Funding; Trillium: Research Funding; Seattle Genetics: Research Funding; Affimed: Research Funding; Bristol-Myers Squibb: Research Funding; Regeneron: Research Funding; LAM Therapeutics: Research Funding; LAM Therapeutics: Research Funding; Seattle Genetics: Research Funding; Affimed: Research Funding; Regeneron: Research Funding; Bristol-Myers Squibb: Research Funding; Bristol-Myers Squibb: Research Funding; Seattle Genetics: Research Funding; Trillium: Research Funding; Seattle Genetics: Research Funding; Trillium: Research Funding; LAM Therapeutics: Research Funding; Affimed: Research Funding; Mayo Clinic Rochester: Employment; Mayo Clinic Rochester: Employment; Mayo Clinic Rochester: Employment; Regeneron: Research Funding; Affimed: Research Funding; Seattle Genetics: Research Funding; Affimed: Research Funding; Trillium: Research Funding; Bristol-Myers Squibb: Research Funding; Seattle Genetics: Research Funding; Regeneron: Research Funding; Bristol-Myers Squibb: Research Funding; LAM Therapeutics: Research Funding; LAM Therapeutics: Research Funding; Mayo Clinic Rochester: Employment; Mayo Clinic Rochester: Employment; Regeneron: Research Funding; Regeneron: Research Funding; Trillium: Research Funding; Mayo Clinic Rochester: Employment; Trillium: Research Funding; Affimed: Research Funding; Bristol-Myers Squibb: Research Funding; Mayo Clinic Rochester: Employment; Affimed: Research Funding; Bristol-Myers Squibb: Research Funding; Affimed: Research Funding; Seattle Genetics: Research Funding; Trillium: Research Funding; LAM Therapeutics: Research Funding; Affimed: Research Funding; Seattle Genetics: Research Funding; Regeneron: Research Funding; Trillium: Research Funding; Trillium: Research Funding; Bristol-Myers Squibb: Research Funding; LAM Therapeutics: Research Funding; Mayo Clinic Rochester: Employment; LAM Therapeutics: Research Funding; LAM Therapeutics: Research Funding; Mayo Clinic Rochester: Employment. Cerhan:Janssen: Membership on an entity's Board of Directors or advisory committees; Celgene: Research Funding; NanoString: Research Funding. Novak:Celgene Coorperation: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal