The bone marrow (BM) microenvironment is increasingly recognized as an important contributor to acute myeloid leukemia (AML) pathogenesis. However, despite growing interest in characterizing different components and cellular architecture of the BM niche and their biological significance in leukemogenesis, the proteomic constitution of the BM extracellular compartment that distinguishes a leukemic niche from its normal counterpart has not yet been fully described.
We therefore performed a quantitative, large-scale proteomic analysis of 1,305 human proteins of the non-cellular compartment of BM (plasma) samples from ten relapsed or refractory AML patients and from ten age- and sex-matched healthy donors (HDs) using an aptamer-based, highly multiplexed, affinity proteomics platform (SOMAscan). This screen identified a total of 168 differentially abundant proteins, of which 91 were significantly more and 77 proteins significantly less abundant in leukemic BM compared with healthy marrow (FC ≥ 1.5, FDR ≤ 0.05). Comparative analysis of BM plasma and peripheral blood (PB) serum samples from the same AML patients and HDs revealed 65 similarly regulated proteins (37 up-regulated vs. 28 down-regulated) and 1 differently regulated protein between the two compartments. Out of the total 168 proteins, 102 proteins were specifically dysregulated only in the BM compartment.
TruSeq Stranded Total RNA-sequencing (Illumina) was also performed using paired-end 75bp sequencing on a HiSeq 3000. RNA was isolated from PAXgene BM RNA tubes (Qiagen) collected in parallel with samples for proteomic analysis. Results of analysis of differentially expressed transcripts only partially overlapped with those candidates identified from our validated proteomic approach, indicating that sequencing of RNA derived from cellular sources of BM may be a suboptimal screening strategy to determine the true proteomic composition of the extracellular compartment of the AML marrow microenvironment.
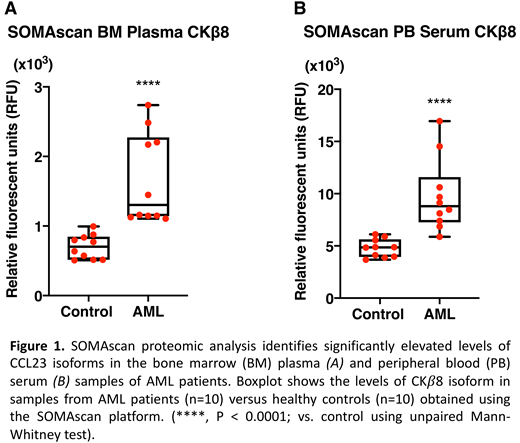
In addition to several previously reported proteins, our proteomics screen discovered numerous aberrantly expressed proteins in leukemic marrow whose role in AML pathogenesis is currently unknown. Using pathway analysis, we identified sets of proteins enriched for specific biological pathways including RAS, ephrin, PDGF, PI3K/AKT, MAPK, Notch, TLR, JAK-STAT, NFκB, Rap1, and Tie2 signaling pathways. A systems biology analysis approach revealed the highly connected network of cytokines and chemokines as the most striking AML-associated proteomic alteration in the BM. We identified IL-8 as a differentially expressed and key central molecule of this network in AML, consistent with recent reports. Importantly, we also identified significantly elevated levels of CKβ8 and CKβ8-1, alternatively spliced isoforms of the myelosuppressive chemokine CCL23 also known as myeloid progenitor inhibitory factor 1 (MPIF-1) or CKβ8, in both leukemic marrow and PB serum samples (Figure 1). Given the critical importance of cytopenias, often disproportional to the degree of leukemic marrow involvement, in the morbidity and mortality of patients with myelodysplastic syndrome (MDS) and AML, we subsequently confirmed this striking finding by performing orthogonal validation in a larger cohort of MDS and AML patients using an ELISA-based immunoassay. This novel finding suggests the possibility that CCL23 may play a role in suppression of normal hematopoiesis in MDS and AML. In support of this hypothesis, we demonstrated in vitro myelosuppressive effects of CCL23 isoforms on colony formation by human CD34+ hematopoietic stem and progenitor cells (HSPCs) in an in vitro colony forming unit assay, resulting in an approximately 2.5-fold decrease in CFU-GM and an evident decrease in CFU-GEMM counts.
In summary, our broad and quantitative proteomic dataset of extracellular factors present in leukemic and normal aging bone marrow has already provided novel mechanistic insights into AML pathogenesis and should serve, together with paired RNA-sequencing information, as a useful public resource for the research community.
Lai:Jazz Pharma: Membership on an entity's Board of Directors or advisory committees; Jazz Pharma: Speakers Bureau; Astellas: Speakers Bureau; Daiichi-Sankyo: Membership on an entity's Board of Directors or advisory committees; Agios: Membership on an entity's Board of Directors or advisory committees. Hourigan:SELLAS Life Sciences Group AG: Research Funding; Merck, Sharpe & Dohme: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal