Background: In multicolor flow cytometry (MFC), switching to a device that supports more fluorochromes per measurement is a common transition in a diagnostic laboratory. Usually this process involves a period of applying both protocols in parallel, the old one validating the new one. Mostly, only very few of the rare diagnoses will be assessed by both protocols. With respect to decision support systems that are based on artificial intelligence this is too little data to train a new classifier considering the need for high accuracy. New approaches of applying existing knowledge to new settings are therefore desirable.
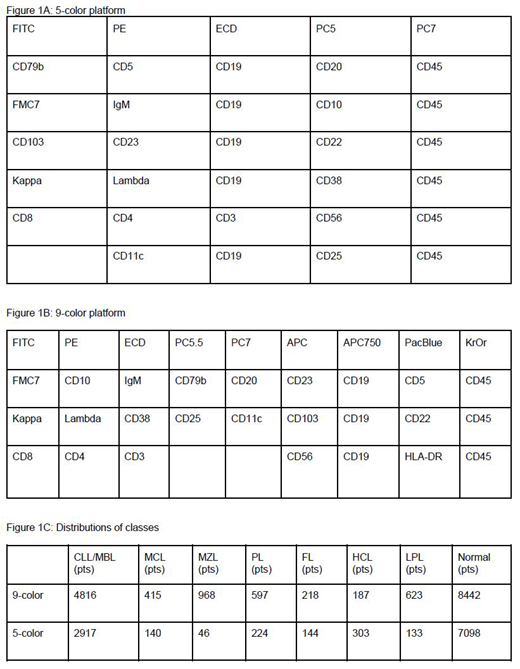
Methods: We obtained MFC data (fcs files) for the two separate sets of samples that were analyzed for the same markers but applying different fluorochrome conjugates and different combinations of antibodies as either 5-color or 9-color combinations both analyzed on the same cytometer. The former platform consisted of 6 combinations with the markers shown in Figure 1A. The latter platform consisted of 3 combinations with the markers shown in Figure 1B. Samples were assigned to one of the B-cell neoplasm classes CLL/MBL, mantle cell lymphoma (MCL), CLL/PL (PL), lymphoplasmacytic lymphoma (LPL), marginal zone lymphoma (MZL), follicular lymphoma (FL), hairy cell leukemia (HCL) or to normal.
We have applied transfer learning (TL), which is a machine learning method to improve the learning of a new task through the transfer of knowledge from a related task that has already been learned. We first train a base network on a base dataset and task, and then we transfer the learned features to a second target network to be trained on a separate dataset. In the present study TL was applied to two different types of MFC protocols. We trained a multi-layer neural network for each of the MFC protocols, while the output layer of both networks remained the same, that is the assignment of unclassified MFC data to seven subtypes of B-cell neoplasms or to the class of absence of B-cell neoplasm. However, for the two different protocols we modified the lower layers of the network to fit the respective combinations of analyzed antigens. We initialized the weights of the upper 5 layers in the 5-color network consisting of a dense layer with 128 nodes, a batch normalization layer with 128 nodes, a second dense layer with 64 nodes, a batch normalization layer with 64 nodes and a final dense layer with 8 nodes with weights from the same layers in the 9-color network. The weight matrices for the dense layers are of the form (wi, bi) where the first component is the weights learnt and the second is the bias vector, each of these vectors of the shape (number of nodes in previous layer * number of nodes in the current layer).
Results: We first assessed the performance of the classification processes for networks that were solely trained on the MFC data of one of the two sets. The distributions of classes for both sets were as shown in Figure 1C. For a random 50/50 split into training and test samples of this data we achieved an overall accuracy of 0.70 for the 5-color data, compared to a 0.82 accuracy for the 9-color data. When we adjusted for the same number of samples in each subtype, there were no significant differences between the two sets, proving the equivalence of both protocols. To assess the benefit of transfer learning, we initialized the upper layers of the 5-color model with the weights of the 9-color model, as more data was in total available from this data set. With knowledge transfer we already achieved an accuracy of 0.7 with 1000 training samples and an accuracy of 0.82 with 4000 training samples. Even more prominent was the improvement of the performance for the rare subtypes, FL, HCL, LPL, PL, MCL and MZL. With the training set consisting of 400 samples from the rare subtypes, the accuracy obtained for these classes was only 0.15; with transfer learning the accuracy for these rare subtypes increased by two thirds. The most prominent improvement of specificity was seen for the subtype FL from 0.35 to 0.57.
Conclusion: We created a framework that enables knowledge transfer for the interpretation of MFC data in B-cell neoplasm diagnostics by an artificial neural network. Our approach is not only applicable for transitions to new protocols, it also allows pooling data of different sources. In so doing, the accuracy for the detection of rare B-cell neoplasm subtypes can be increased by joining forces in a collaborative effort by involving multiple different data sets.
Elsner:res mechanica: Employment, Equity Ownership. Lueling:res mechanica: Employment, Equity Ownership. Schabath:MLL Munich Leukemia Laboratory: Employment. Haferlach:MLL Munich Leukemia Laboratory: Employment, Equity Ownership. Haferlach:MLL Munich Leukemia Laboratory: Employment, Equity Ownership. Kern:MLL Munich Leukemia Laboratory: Employment, Equity Ownership.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal