Introduction: AML is a heterogeneous disease with diverse patient outcomes. During the last 4 decades, several cytogenetic and molecular markers have been used for risk stratification of AML pts and to guide therapeutic decisions. In 2017, Gerstung et al. (Nat Genet 2017;49:332) used a knowledge bank (KB; i.e., combination of clinical, outcome, cytogenetic and sequencing data of 111 genes from 1,540 AML pts) to generate an algorithm that is able to predict likelihoods for remission, relapse, and mortality in AML pts. The prognostic value of the established KB algorithm was validated in two independent but smaller pt cohorts (Gerstung et al.: n=186 pts; Huet et al. Blood 2018;132:865: n=155 pts).
Aims: The aim of our study was to validate the prognostic relevance of the KB algorithm both in our entire large independent adult pt cohort and in age subgroups (i.e., younger [<60 y] and older [≥60 y] pts). We also compared the performance of the KB algorithm with that of another well-established prognostic classification, the 2017 European LeukemiaNet (ELN) genetic risk classification (Döhner et al. Blood 2017;129:424).
Methods: We analyzed 1,617 pts (median age: 53 y; 1,048 aged <60 y and 569 aged ≥60 y; including 167 early death [ED] pts who died within 30 days after diagnosis) with de novo AML appropriate for intensive chemotherapy. The mutational status of 80 cancer- and leukemia-associated genes (Eisfeld et al. Leukemia 2017;31:2211) was determined using a targeted next-generation sequencing panel (variants with variant allele fraction < 5% were defined as not mutated) The status of biallelic CEBPA mutations was determined using Sanger sequencing, and an internal tandem duplication of the FLT3 gene using fragment analysis in pretreatment bone marrow or blood samples. All pts were treated on CALGB/Alliance protocols and none received an allogeneic transplant in first complete remission (CR1). Of the genes whose mutational status is included in the KB algorithm, 26% were not included in our sequencing panel, but all 30 genes that had the biggest impact on the algorithm's ability to predict outcome were contained in our panel.
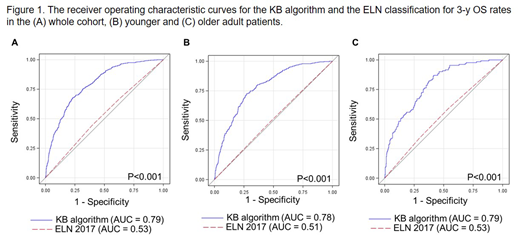
Results: We used the 3-y overall survival (OS) rates to compare the KB algorithm prediction with the actual outcome. In the whole cohort, we found the area under the receiver operating characteristic curve (AUC) to be AUCKB = 0.79 (Figure 1A). Of note, AUC = 1.00 means perfect prediction ability whereas AUC = 0.50 denotes lack of prediction ability equal to that of random chance. Concerning other clinical endpoints, we found that the KB approach had the highest AUC for predicting non-remission death (i.e., pts who died without achieving a CR1, AUCKB = 0.84), followed by relapse death (i.e., pts who died after relapse, AUCKB = 0.69), and non-relapse death (i.e., pts who died in CR1, AUCKB = 0.61). Analysis of the 3-y OS in the subgroup of younger pts yielded similar results with an AUCKB = 0.78. Older pts are known to have poorer prognosis and risk stratification is more difficult for this cohort, but, we still found an AUCKB = 0.79 for the KB approach.
Next, we compared the predictive value of the KB approach with the current ELN classification and found that KB outperformed the ELN classification in the whole cohort (AUCKB = 0.79 vs AUCELN = 0.53, P<0.001; Figure 1A) as well as in the younger (AUCKB = 0.78 vs AUCELN = 0.51, P<0.001; Figure 1B) and older pt subgroups (AUCKB = 0.79 vs AUCELN = 0.53, P<0.001; Figure 1C). We also compared KB and ELN after excluding ED pts because the ELN risk group is a poor predictor for ED. We found that KB still outperformed the ELN classification in the whole cohort (AUCKB = 0.78 vs AUCELN = 0.71, P<0.001) and in the younger pts (AUCKB = 0.74 vs AUCELN = 0.69, P=0.005). However, in older pts the difference was of borderline significance (AUCKB = 0.75 vs AUCELN = 0.70, P=0.05).
Conclusions: Our analysis of a large cohort of 1,617 pts with de novo AML treated with intensive chemotherapy validated the prognostic value of the recently published KB algorithm for the 3-y OS endpoint. Although we found that the KB approach had a high predictive relevance for non-remission death, the AUCs for relapse death and non-relapse death were lower. We also showed that the KB approach had a better predictive value than the current ELN classification but the differences in the AUCs were smaller when ED pts were excluded.
Support: CA233338, U24CA196171, U10CA180821, U10CA180882.
https://acknowledgments.alliancefound.org
Kolitz:Boeringer-Ingelheim: Research Funding; Astellas: Research Funding; Roche: Research Funding. Powell:Jazz Pharmaceuticals: Consultancy, Research Funding, Speakers Bureau; Pfizer: Consultancy, Research Funding; Rafael Pharmaceuticals: Consultancy, Research Funding; Novartis: Consultancy, Speakers Bureau; Janssen: Research Funding. Stone:AbbVie, Actinium, Agios, Argenx, Arog, Astellas, AstraZeneca, Biolinerx, Celgene, Cornerstone Biopharma, Fujifilm, Jazz Pharmaceuticals, Amgen, Ono, Orsenix, Otsuka, Merck, Novartis, Pfizer, Sumitomo, Trovagene: Consultancy; Argenx, Celgene, Takeda Oncology: Other: Data and Safety Monitoring Board/Committee: ; Novartis, Agios, Arog: Research Funding. Byrd:Janssen: Consultancy, Other: Travel Expenses, Research Funding, Speakers Bureau; Gilead: Other: Travel Expenses, Research Funding, Speakers Bureau; Ohio State University: Patents & Royalties: OSU-2S; Pharmacyclics LLC, an AbbVie Company: Other: Travel Expenses, Research Funding, Speakers Bureau; Ohio State University: Patents & Royalties: OSU-2S; Ohio State University: Patents & Royalties: OSU-2S; Acerta: Research Funding; TG Therapeutics: Other: Travel Expenses, Research Funding, Speakers Bureau; Acerta: Research Funding; Pharmacyclics LLC, an AbbVie Company: Other: Travel Expenses, Research Funding, Speakers Bureau; Pharmacyclics LLC, an AbbVie Company: Other: Travel Expenses, Research Funding, Speakers Bureau; TG Therapeutics: Other: Travel Expenses, Research Funding, Speakers Bureau; Novartis: Other: Travel Expenses, Speakers Bureau; TG Therapeutics: Other: Travel Expenses, Research Funding, Speakers Bureau; Genentech: Research Funding; Genentech: Research Funding; Genentech: Research Funding; BeiGene: Research Funding; Novartis: Other: Travel Expenses, Speakers Bureau; Gilead: Other: Travel Expenses, Research Funding, Speakers Bureau; Gilead: Other: Travel Expenses, Research Funding, Speakers Bureau; BeiGene: Research Funding; Janssen: Consultancy, Other: Travel Expenses, Research Funding, Speakers Bureau; Acerta: Research Funding; BeiGene: Research Funding; Janssen: Consultancy, Other: Travel Expenses, Research Funding, Speakers Bureau; Novartis: Other: Travel Expenses, Speakers Bureau.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal