The transthyretin (TTR) and immunoglobulin light chain proteins do not exist in isolation in cardiac amyloid (CA) plaques. Laser microdissection followed by mass spectrometry-based proteomic (LMD-MS) typing techniques can identify hundreds of co-deposited proteins in the "nano-environment" of amyloid deposits. Some, such as the "amyloid signature" proteins (SAP, APOE, APOA4, vitronectin, clusterin), are specific for amyloid plaques across different tissues and amyloid types. However, many of the remaining proteins are also abundant in the serum or normal tissues making it difficult to discern if they are unique to the amyloid plaque or part of the normal tissue background.
We included 288 patients/samples with TTR CA (of which 53 were mutated) diagnosed by endomyocardial biopsy and 5 normal control samples obtained from 3 patients during autopsy. At the time of the meeting updated analyses including 168 cardiac light chain amyloid patients and 15 controls from patients with restrictive and hypertensive cardiomyopathy will be presented. Normalized spectral counts were used as a semi-quantitative measure of abundance. Proteins were considered part of the proteome if their abundance in the amyloid plaque was increased by 1.5-fold compared to normal controls (FDR p<0.05).
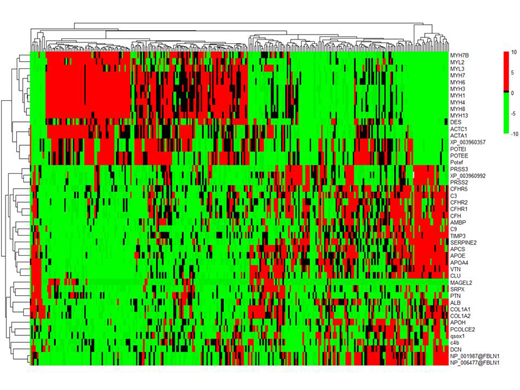
Of 2240 unique proteins identified across all samples, 42 were differentially expressed in amyloid plaques. These included a) signature proteins: SAP, APOE, APOA4, vitronectin, clusterin; b) serum proteins: albumin, b2-glycoprotein, 7 complement pathway proteins; c) matrix proteins: 2 collagen isoforms, fibulin 1, decorin, PCOLCE2, TIMP3; d) cytosketal and contractile proteins: 2 actin isoforms, MYL2, 8 myosin isoforms, 3 ankyrin isofroms, desmin; and "other" proteins: serpine2, MAGE-like protein-2, pleiotrophin, AMBP, SRPX and QSOX1). The signature and cytoskeletal proteins were the most abundant overall, however, proteins that were most increased compared to normal included (log2 fold change in parentheses): fibulin (24.5), PCOLCE2 (23.5), b2-glycoprotein (23.5), factor H related peptide-2 (24.2), serpine2 (24.7), mage-like protein-2 (22.8), pleiotrophin (23.8), QSOX1 (22.9), SRPX (23.5). B2 glycoprotein was more abundant in wild type TTR samples(p=0.003). APOE (p=0.002), fibulin (p=0.008) and serpine2 (p<0.0001) were more abundant in UK stage (Gilmore JD et al, Eur Heart J 2018) 2/3 patients. Unsupervised hierarchical clustering of patients by normalized protein abundance revealed two major distinct patterns of co-deposited proteins (figure). Patients with high levels of contractility proteins (myosin isoforms, desmin, actins) were characterized by low levels of most other proteins (clusters on the left). Most complement proteins co-deposited along with higher levels of signature proteins (clusters on the right). When comparing clinical and laboratory characteristics between those 2 groups of patients only cardiac strain was found to be significantly higher (worse) in patients with lower levels of contractility proteins (p=0.01).
Our data shows that the TTR CA proteome includes signature, serum and cardiac tissue proteins with variable expression according to TTR mutation status and cardiac functional status, which could inform mechanisms of disease pathogenesis. Comparison with diseased controls may further clarify if this proteomic pattern is specific to CA.
Dasari:The Binding Site: Patents & Royalties: US Patent Rights on Mass Spectroscopy Licensing agreement with The Binding Site, Research Funding. Dispenzieri:Akcea: Consultancy; Intellia: Consultancy; Janssen: Consultancy; Pfizer: Research Funding; Takeda: Research Funding; Celgene: Research Funding; Alnylam: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal