Introduction: The clinical presentation and frequency of organ involvement are quite distinct in patients with IgM amyloidosis compared to patients with non-IgM amyloidosis, with less heart involvement and more nerve, soft tissue and lung involvement in IgM patients. It has been observed that immunoglobulin light chain variable region (IGVL) gene and gene family of the plasma cell clone impact phenotypic manifestations in AL amyloidosis, particularly organ tropism. (Kourelis et al. Blood. 2017;129). Given the variability in clinical features amongst patients with IgM vs. non IgM AL amyloidosis, we hypothesized that IGVL gene usage would be different between these two groups.
Methods: Patients with newly diagnosed IgM and non-IgM amyloidosis seen at our institution from 01/2006 to 12/2015 were identified. IGVL gene usage was assessed by liquid chromatography tandem mass spectrometry (LC/MS/MS) as previously described. (Vrana et al Blood 2009;Dasari et al J of Proteome Res; Kourelis et al. Blood. 2017). Mass spectrometry data for IGVL analysis was obtained from clinically available data. LC/MS/MS was performed on available archival specimens when such data was not available clinically.
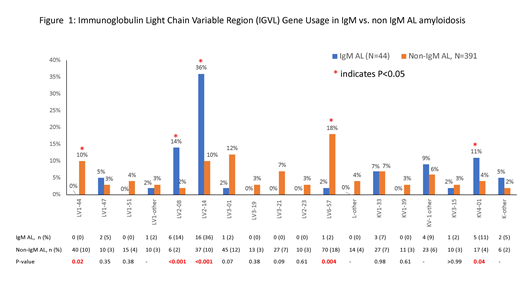
Results: Patients with newly diagnosed amyloidosis who had sufficient mass spectrometry data on fat aspirate or tissue biopsy for IGVL gene usage identification were included (IgM AL, N=44 and Non-IgM AL, N=391).
Clinical Features: As expected, differences were noted in baseline characteristics of patients with IgM vs. non-IgM AL amyloidosis. Patients with IgM amyloidosis were older (68 vs. 64 years, p=0.04) and more likely to have kappa as the involved light chain (34% vs. 25%, p=0.2), though the difference did not reach statistical significance. IgM amyloidosis patients had lower difference in involved and uninvolved free light chains (dFLC) (15.5 vs. 25.8 mg/dL, p=0.01) and lower NTProBNP levels (median: 1987 vs. 2655 pg/mL, p=0.02) compared with non-IgM amyloid patients. Presence of t(11;14) was less common by FISH (30% vs. 48%, p=0.1), though the difference did not reach statistical significance. Patterns of organ involvement were different, with heart involvement being less frequent in IgM patients (64% vs. 76%), while peripheral nerve (30% vs. 18%, p=0.05), soft tissue (41% vs. 21%, p=0.005) and lung involvement (7% vs. 3%, p=0.1) were more common in patients with IgM vs. non-IgM AL amyloidosis. There was no difference in liver (9% vs. 15%), kidney (50% vs. 51%) GI (23% vs. 25%) or autonomic nervous system involvement (18% vs. 14%).
IGVL Gene Usage: As shown in Figure 1, IGVL usage differed across the two groups. In the lambda group, LV2-08 (14% vs. 2%, p<0.001) and LV2-14 (36% vs. 10%, p<0.001) were more common in IgM amyloid patients, while LV1-44 (0% vs. 10%, p=0.02) and LV6-57 (2% vs. 18%, p=0.004) were less common in the IgM group. There was also a trend towards less frequent involvement of LV3-01 (2% vs. 12%, p=0.07) in IgM amyloid patients. In the kappa group, KV4-01 (11% vs. 4%, p=0.04) was more common in patients with IgM amyloidosis.
Correlation Between IGVL Gene usage and Clinical Features: IGVL gene usage correlated with disease characteristics and differences in the clinical presentation of patients across the two groups. LV2-14 was more commonly seen in IgM amyloid patients (above). It has been previously associated with a higher frequency of peripheral nerve involvement (Kourelis et al. Blood 2017), which was more frequent in IgM group. LV2-14 is also associated with low dFLC levels, which were noted to be lower in the IgM AL cohort. LV1-44 has been associated with higher likelihood of cardiac involvement. LV1-44 was seen less frequently in the IgM amyloid patients, who also had lower rates of cardiac involvement. LV6-57 has been historically associated with higher rates of t(11;14). It was less common in IgM AL patients, who also had lower rates of t(11;14).
Conclusions: IGVL gene usage differed significantly in patients with IgM vs. non-IgM amyloidosis, correlating with differences in disease characteristics and organ involvement. This is the first study to our knowledge to report IGVL gene usage differences in IgM vs. non-IgM AL amyloidosis. Differences in IGVL gene usage may explain the distinct clinical presentation seen in IgM amyloidosis, the biology of which has so far remained poorly understood.
*SS and SD contributed equally
Dasari:The Binding Site: Patents & Royalties: US Patent Rights on Mass Spectroscopy Licensing agreement with The Binding Site, Research Funding. Dispenzieri:Alnylam: Research Funding; Celgene: Research Funding; Takeda: Research Funding; Pfizer: Research Funding; Janssen: Consultancy; Akcea: Consultancy; Intellia: Consultancy. Murray:The Binding Site: Patents & Royalties: US Patent Rights on Mass Spectroscopy Licensing agreement with The Binding Site, Research Funding. Kumar:Janssen: Consultancy, Research Funding; Celgene: Consultancy, Research Funding; Takeda: Research Funding. Gertz:Prothena: Honoraria; Alnylam: Honoraria; Ionis: Honoraria; Janssen: Honoraria; Spectrum: Honoraria, Research Funding; Celgene: Honoraria.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal