Introduction
Non-Hodgkin B-cell lymphomas (B-NHLs) are a highly heterogeneous group of mature B-cell malignancies associated with very diverse clinical behaviors. They rely on the activation of different signaling pathways for proliferation and survival which might be amenable to targeted therapies, increasing the need for precision diagnosis. Unfortunately, their accurate classification can be challenging, even for expert hemato-pathologists, and secondary reviews recurrently differ from initial diagnosis. To address this issue we have developed a pan-B-NHL classifier based on a middle throughput gene expression assay coupled with a random forest algorithm.
Material and Methods
Five hundred ten B-NHL diagnosed according to the WHO criteria were studied, with 325 diffuse large B-cell lymphomas (DLBCL), 43 primary mediastinal B-cell lymphomas (PMBL), 55 follicular lymphomas (FL), 31 mantle cell lymphomas (MCL), 17 small lymphocytic lymphomas (SLL), 20 marginal zone lymphomas (MZL), 11 marginal zone lymphomas of mucosa-associated lymphoid tissue (MALT) and 8 lymphoplasmacytic lymphomas (LPL). To train and validate the predictor the samples were randomly split into a training (2/3) and an independent validation cohort (1/3). A panel of 137 genes was designed by purposely selecting the differentiation markers identified in the WHO classification for their capacity to provide diagnostic and prognostic information in NHLs. Gene expression profiles were generated by ligation dependent RT-PCR applied to RNA extracted from frozen or FFPE tissue and analyzed on a MiSeq sequencer. For analysis, the sequencing reads were de-multiplexed, aligned with the sequences of the LD-RTPCR probes and counted. Results were normalized using unique molecular indexes counts to correct PCR amplification biases.
Results
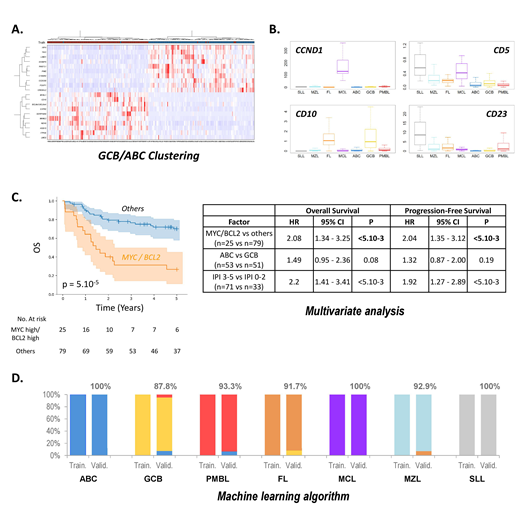
In DLBCL, unsupervised gene expression analysis retrieved the expected GCB, ABC and PMBL signatures (Fig A). These tumors also showed higher expressions of the KI67 (proliferation), CD68 and CD163 (tumor associated macrophages), and PD-L1/2 (immune response) markers. We also observed that the dual expression of MYC and BCL2 at the mRNA level significantly associates with inferior PFS and OS, independent from the International Prognostic Index and from the GCB/ABC cell-of-origin signature, validating the capacity of the assay to identify these highly aggressive lymphomas (Fig C).
Overall, low-grade lymphomas were characterized by a significant T cell component. FLs associated with the GCB (BCL6, MYBL1, CD10 and LMO2) and Tfh (CD3, CD5, CD28, ICOS, CD40L, CXCL13) signatures. Other small B-cell lymphomas tended to overexpress activated B-cell markers (LIMD1, TACI, IRF4,FOXP1...), and the expected CD5, CD10, CD23 and CCND1 differential expressions in SLL, MCL and MZL were correctly retrieved (Fig B). Surprisingly, our analysis revealed that the Ie-Ce sterile transcript, expressed from the IGH locus during IgE isotype switching, is almost exclusively expressed by FLs, constituting one of the most discriminant markers for this pathology.
We next trained a random forest classifier to discriminate the 7 principal subtypes of B-NHLs. The training cohort comprised 162 DLBCLs (ABC or GCB), 28 PMBL, 35 FLs (grade 1-3A), 21 MCLs, 12 SLLs, and 25 NHLs grouped into the MZL category (13 MZLs, 8 MALT and 4 LPLs). The independent validation series comprised 90 DLBCLs classified as GCB or ABC DLBCLs by the Lymph2Cx assay, 15 PMBLs, 12 FLs (grade 1-3A), 10 MCLs, 5 SLLs and 14 MZLs (7 MZL, 3 MALT and 4 LPL). The RF algorithm classified all cases of the training series into the expected subtype, as well as 94.5% samples of the independent validation cohort (Fig D). For ABC and GCB DLBCLs, the concordance with the Lymph2Cx assay in the validation cohort was 94.3%.
Conclusion
We have developed a comprehensive gene expression based solution which allows a systematic evaluation of multiple diagnostic and prognostic markers expressed by the tumor and by the microenvironment in B-NHLs. This assay, which does not require any specific platform, could be implemented in complement to histology in many diagnostic laboratories and, with the current development of targeted therapies, enable a more accurate and standardized B-NHL diagnosis. Together, our data illustrate how the integration of gene expression profiling and artificial intelligence can increase precision diagnosis in cancers.
Oberic:Takeda: Membership on an entity's Board of Directors or advisory committees; Janssen: Honoraria; Roche: Membership on an entity's Board of Directors or advisory committees. Haioun:Miltenyi: Honoraria; Takeda: Honoraria; Servier: Honoraria; F. Hoffmann-La Roche Ltd: Honoraria; Novartis: Honoraria; Amgen: Honoraria; Celgene: Honoraria; Gilead: Honoraria; Janssen: Honoraria. Salles:Roche, Janssen, Gilead, Celgene: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Other: Educational events; Amgen: Honoraria, Other: Educational events; BMS: Honoraria; Merck: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees; Novartis, Servier, AbbVie, Karyopharm, Kite, MorphoSys: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Other: Educational events; Autolus: Consultancy, Membership on an entity's Board of Directors or advisory committees; Takeda: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Other: Educational events; Epizyme: Consultancy, Honoraria. Tilly:roche: Membership on an entity's Board of Directors or advisory committees; servier: Honoraria; merck: Honoraria; Roche: Consultancy; Celgene: Consultancy, Research Funding; Astra-Zeneca: Consultancy; Karyopharm: Consultancy; BMS: Honoraria; Janssen: Honoraria; Gilead: Honoraria. Jardin:celgene: Honoraria; roche: Honoraria; amgen: Honoraria; Servier: Honoraria; janssen: Honoraria.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal