Background:
Mutations in the nucleophosmin (NPM1) gene are associated with better responses to chemotherapy and improved survival among acute myeloid leukemia (AML) patients. However, older AML patients (≥ 60 years old) with NPM1 mutation have worse survival outcomes than younger patients (<60 years old). This may be attributed to more adverse biologic features (frequent complex karyotype, FLT3 mutations) in addition to lower odds to receive intensive curative chemotherapy due to co-morbidities. We sought to compare the outcomes of older NPM1 mutated AML patients with younger NPM1 mutated patients after exclusions of patients with adverse-risk per ELN 2017 criteria. We also compared their genomic mutation profile and gene expression utilizing the Beat AML dataset.
Methods:
We queried the Beat AML dataset, supported in part by the Leukemia & Lymphoma Society and the OHSU Knight Cancer Institute, for pts with NPM1 gene mutations who did not have adverse-risk ELN 2017 (poor cytogenetic profile or mutations in FLT3, TP53 or ASXL1). Descriptive statistics described baseline characteristics and responses. Kaplan-Meier with log-rank test was used for survival analysis. DNA mutation data were obtained from the exome sequencing and analyzed using the beat AML data viewer (Vizome). RNA exome sequencing data were downloaded. Differential expression of raw count RNA-Seq and gene set enrichment was done using R via limma and ClusterProfiler packages.
Results:
Among 562 unique patients in the Beat AML umbrella trial, there were 81 patients with newly diagnosed NPM1 mutated AML after exclusion of patients with ELN 2017 adverse-risk category. Among these patients there was 49 older patients (≥ 60 years old) and 32 younger patients (<60 years old). 39 (77.6%) in the older group received intensive induction chemotherapy and 30 patients (93.7%) in the younger group. 29 (59.1%) patients achieved complete morphologic responses in the older patient group compared to 28 (84.4%) in the younger patient group (OR 0.2, P=0.009). Median overall survival in the older patient group was 20.1 months compared to 25.4 months in the younger group (HR 0.52, P=0.08).
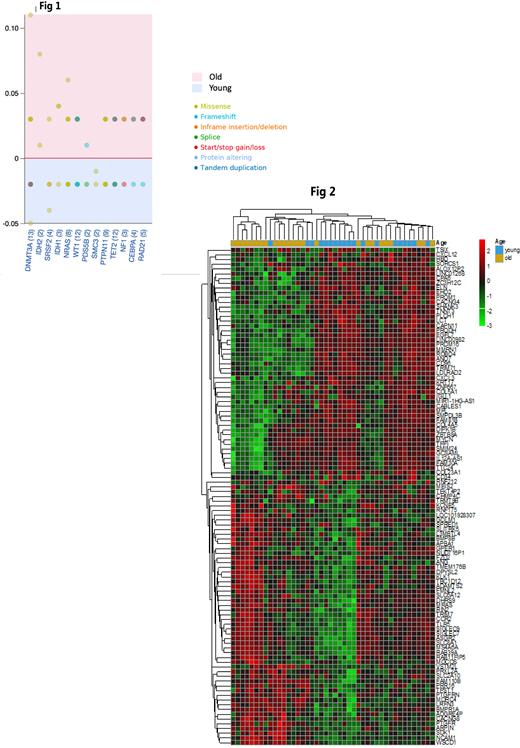
Exome sequencing data were available for 43 and 30 patients from the older and younger group respectively. There was a median of 6.5 (2-20) and 7 (2-19) mutations in the older and younger group respectively (P=0.78). After exclusion of the benign mutations and variant of unknown significance, the median number of mutations was 4 in both group (P=0.28). Both groups shared only 24 (3.9%) of the gene mutations while there were 334 unique gene mutations in the older group and 262 in the younger group. Most common gene mutations were DNMT3a, TET2, NRAS, WT1, and PTPN11 with frequencies are shown (Figure 1). RNA sequencing data was available for 26 patients from the older group and 18 patients from the younger group. We explored the gene expression profile of the top 1000 differentially expressed genes in both groups after adjustment. There was distinctive clustering of the gene expression profile between the two groups (Figure 2). Gene set enrichment analysis identified multiple immune-related pathways among the highly enriched gene sets in both groups but with different functions in the two groups. There was significant gene set enrichment in the TGFβ signaling in the older patient group which is associated with immune suppression and microenvironment modulation. While the younger group showed significant enrichment in the TNFa, IL17, PI3K-AKT signaling which are associated with inflammation.
Conclusion:
Older AML patients with NPM1 mutations, and no adverse risk features, had lower rate of complete responses and a trend towards a worse survival compared to younger patients. Whole exome sequencing did not show increased mutational burden. However, 96% of the mutated genes were different between the two groups as were the gene expression profiles. Gene set enrichment analysis showed contrasting enriched immune-related pathways between both groups. The immunosuppressive TGFβ signaling gene set were significantly enriched in the older group while the inflammatory TNFa, IL17, PI3K-AKT signaling gene sets were significantly enriched in the younger group. Older AML patient with NPM1 mutations have distinctive genomic landscape compared to the younger patient which may explain in part the worse clinical outcomes in the absence of other adverse risk features.
Lin:Jazz Pharmaceuticals: Honoraria; Pfizer: Membership on an entity's Board of Directors or advisory committees.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal