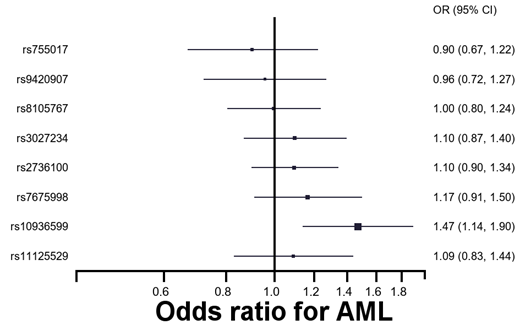
Background Maintenance of telomere length has long been established to play a role in the biology of cancer, and several lines of evidence suggest that it may be especially important in myeloid malignancy. Measuring leukocyte telomere length (LTL) directly in case control studies is problematic because telomere length is likely to be influenced by the disease process. Recent studies have led to the identification of a robust genetic predictor of LTL, which we have used to compare differences in predicted telomere length in case control studies of myeloid leukemia and myelodysplastic syndromes (MDS) using Mendelian randomization (MR). MR is a method used to measure the association between a trait (e.g. telomere length) and an outcome (e.g. myeloid malignancy) that can't be measured directly by utilizing genetic variants known to be associated with the trait as instrumental variables. The use of genetic predictors of telomere length reduces the possibility that the association is explained by reverse causation and confounding likely to be relevant in case-control studies where telomere length is measured directly because genotypes in the general population should be randomly distributed. Additionally, predicted telomere length has the added advantage of representing telomere length over the lifetime of the individual while measuring LTL directly will represent only one time point in an individual's life. Methods Myeloid leukemia (AML and CML) and MDS cases were identified by rapid case ascertainment through the Minnesota Cancer Surveillance System (MCSS). Centralized pathology and cytogenetics reviews were conducted to confirm diagnosis and classify by subtypes. Controls were identified through the Minnesota State driver's license/identification card list. We used the Sequenom platform to genotype seven SNPs that were validated predictors of LTL in a recently published meta-analysis. We estimated associations between individual SNPs and myeloid malignancy using multivariable logistic regression. Mendelian randomization was used to evaluate the association between predicted LTL and risk of myeloid malignancy. Results We included 253 AML cases, 353 MDS cases, 145 CML cases, and 1,042 controls. We observed a significant association between longer predicted telomere length and AML in analyses adjusted for sex and age (OR 4.82 per additional kilobase of average telomere length, 95% CI 1.32-17.63). The individual SNP analyses suggest that the association is largely driven by rs10936599, located in TERC (OR 1.47 per allele, 95% CI 1.14-1.90; Figure). We did not observe statistically significant associations between predicted LTL and MDS or CML. Discussion In this analysis of predicted telomere length and myeloid malignancy, we identified a significant association between longer predicted leukocyte telomere length and AML. In contrast, no significant association was observed for MDS or CML although we did observe an elevated OR for longer predicted telomere length and MDS that did not reach statistical significance.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal