Background: Human leukocyte antigen (HLA) genes on chromosome 6p21 encode the major histocompatibility complex (MHC). MHC molecules are expressed on all nucleated cells and are essential to the immune system's ability to distinguish healthy native cells from foreign or abnormal ones. Specialized antigen presenting cells display MHC molecules loaded with foreign peptides to initiate T cell responses. HLA sequencing variability is critical to function; studies have shown even single allelic changes can alter the ability of MHC molecules to recognize cognate T cell receptors. Given the role of MHC molecules in identifying cells as "self", associations between HLA sequence variation and autoimmunity is a research focus. Like other autoimmune disorders, association between ITP and HLA genes has been investigated. Prior studies are limited by outdated HLA typing methods, genetically homogenous study populations, and lack of validation studies.
Preexisting exome data has historically not been used to provide accurate HLA sequencing information given the polymorphic, highly repetitive sequence in this region. This study applied a novel bioinformatics pipeline, xHLA, to an existing set of whole exome sequencing in a cohort of chronic pediatric ITP patients. xHLA, reported by Xie et al. in 2017, yields high-resolution, two field HLA typing for HLA-A, -B, -C, -DQB1, -DPB1, and -DRB1.
A prior genetic analysis of chronic ITP patient whole exome data identified a missense variant in BTNL2, a gene which plays a role in peripheral regulatory T cell induction. This variant (rs143211074) is upregulated in chronic ITP versus healthy controls (minor allele frequency= 4.17% in ITP v. 0.54% in controls, p=1.43x10-4). Linkage of rs143211074 and HLA alleles was examined in this study.
Methods: Whole exome sequencing data for a cohort of 272 chronic ITP patients was processed using the xHLA method. In order to eliminate sequencing variability related to ethnicity alone, initial analyses were focused on Caucasians only (n=170), using Eigenvectors for principle components analysis. The frequency of individual HLA alleles and HLA haplotypes for Caucasians from the National Marrow Donor Program (NMDP) Be The Match Registry® were compared to the ITP cohort. HLA haplotypes were inferred using the NMDP HaploStats program. A chi-squared test was utilized to compare nonparametric categorical data using GraphPad Prism version 8.0.1 for Windows, GraphPad Software, San Diego, California, USA, www.graphpad.com. A Bonferroni correction was applied and a p-value of <0.05 was statistically significant.
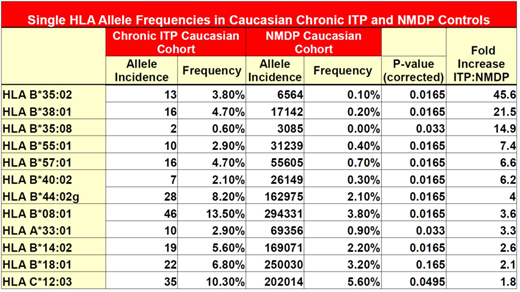
Results: 12 HLA alleles had increased frequency in the chronic ITP cohort versus controls. (Table 1) The BTNL2 variant, rs143211074, was found in linkage disequilibrium with the HLA-DRB1*04:02, HLA-DQB1*03:02 haplotype. Of the 10 patients heterozygous for the variant allele, 80% had the HLA-DRB1*04:02, HLA-DQB1*03:02 haplotype. Both patients homozygous for rs143211074 were also homozygous for the HLA-DRB1*04:02, HLA-DQB1*03:02 haplotype. In the entire cohort, this haplotype was identified in 3.63% of ITP patients, and only in 1.27% of controls.
Our group recently published that a positive direct antiglobulin test is associated with chronic ITP and need for second line treatments. Of the patients with the HLA-DRB1*04:02, HLA-DQB1*03:02 haplotype with DAT testing performed (n=9), 66% had a positive DAT test and 33% had a negative DAT result. Of those without the HLA-DRB1*04:02, HLA-DQB1*03:02 haplotype who had DAT testing, 17.2% had a positive DAT and 82.8% had a negative test (p=0.0026).
Conclusions: HLA alleles are seen at increased frequency in a cohort of Caucasian chronic ITP patients. The HLA-DRB1*04:02, HLA-DQB1*03:02 haplotype is linked to a variant previously identified in BTNL2. Coupled with prior data suggesting that a positive DAT result is associated with chronic disease and use of second line agents, this presence of the HLA-DRB1*04:02, HLA-DQB1*03:02 haplotype may also be linked to refractory disease.
Confirmation of the accuracy of the xHLA method in a cohort of patients with both exome data and traditional HLA typing is underway. In the future, a separate cohort of chronic and acute ITP patients will undergo next generation sequencing and have similar HLA typing performed as an independent validation set.
Despotovic:Novartis: Research Funding; Amgen: Research Funding; Dova: Honoraria.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal