In this issue of Blood, Mottok et al present a large case series of well defined primary mediastinal B-cell lymphoma (PMBL) that includes whole genome sequencing of all 94 cases and molecular confirmation of diagnosis in 73 cases.1,2 The resulting mutation and copy number data are correlated with gene expression signatures, which confirm previous correlations using smaller data sets as well as extend the description of mechanisms of immune evasion and driver mutations. Altogether, this highly annotated and large PMBL case series should provide a rich and definitive data set for genetic research in this disease.
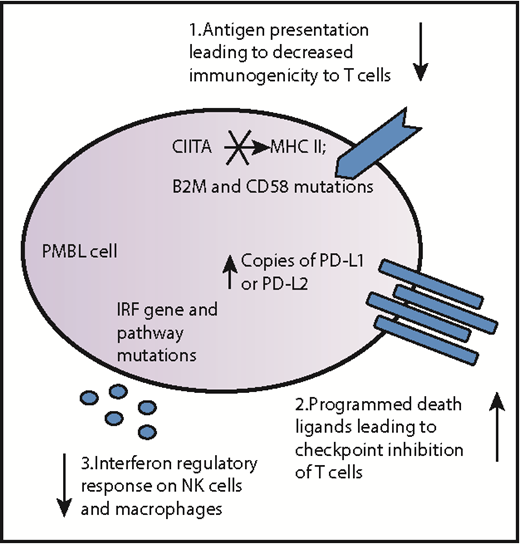
Schematic diagram summarizing immune escape mechanisms of PMBL. PMBL tumors can harbor multiple genetic alterations, including mutations, indels, and copy number alterations, which enable them to evade tumor immunosurveillance. These general approaches to immune evasion firstly include alterations in antigen presentation such as rearrangements, indels, and mutations of the CIITA gene, which encodes the master transactivator of the major histocompatibility complex (MHC) class II molecules; mutations in the gene encoding the invariant chain CD58, also known as the lymphocyte adhesion molecule LFA-3, that in the wild-type state, strengthens the adhesion between the antigen-presenting cell (in this case, the PMBL cell) and reactive T cells; and mutations in the gene encoding β-2 microglobulin (B2M), which in the wild-type state, normally stabilizes the MHC class I molecules to activate CD8+ T cells. Secondly, T-cell activation is affected by amplification of the genes encoding programmed death-ligand 1 (PD-L1) and PD-L2, which bind the inhibitory checkpoint molecule programmed cell death protein 1 (PD-1) on antigen-specific T cells. Thirdly, natural killer (NK) cells and macrophages are affected by mutations in the interferon regulatory response cytokines and downstream genes which, in the wild-type state, upregulate antigen presentation via MHC.
Schematic diagram summarizing immune escape mechanisms of PMBL. PMBL tumors can harbor multiple genetic alterations, including mutations, indels, and copy number alterations, which enable them to evade tumor immunosurveillance. These general approaches to immune evasion firstly include alterations in antigen presentation such as rearrangements, indels, and mutations of the CIITA gene, which encodes the master transactivator of the major histocompatibility complex (MHC) class II molecules; mutations in the gene encoding the invariant chain CD58, also known as the lymphocyte adhesion molecule LFA-3, that in the wild-type state, strengthens the adhesion between the antigen-presenting cell (in this case, the PMBL cell) and reactive T cells; and mutations in the gene encoding β-2 microglobulin (B2M), which in the wild-type state, normally stabilizes the MHC class I molecules to activate CD8+ T cells. Secondly, T-cell activation is affected by amplification of the genes encoding programmed death-ligand 1 (PD-L1) and PD-L2, which bind the inhibitory checkpoint molecule programmed cell death protein 1 (PD-1) on antigen-specific T cells. Thirdly, natural killer (NK) cells and macrophages are affected by mutations in the interferon regulatory response cytokines and downstream genes which, in the wild-type state, upregulate antigen presentation via MHC.
PMBL is an unusual, somewhat enigmatic lymphoma that combines some clinical, cytogenetic, and gene expression profile features of classical Hodgkin lymphoma with the most common aggressive non-Hodgkin B-cell lymphoma, diffuse large B-cell lymphoma (DLBCL).3,4 PMBL is thought to arise from mutations accumulated in thymic B cells and is 1 of only 2 lymphomas, along with nodular sclerosis Hodgkin lymphoma, that are more common in women than men. Other typical clinical features include location in the anterior mediastinum, local tissue invasion, and relatively young patient age at diagnosis. Some data suggest that a more dose-intense treatment regimen than that usually administered for DLBCL may benefit patients with PMBL.5 However, a precise pathologic diagnosis on which to base informative clinical trials can be difficult because PMBL exhibits morphologic and immunophenotypic profiles that overlap with other types of lymphomas and also because morphologically similar DLBCL can occur in the mediastinum whereas PMBL-like tumors can be found at other locations in the body.2,4,6
The study by Mottok et al advances the work on immune evasion in PMBL via (1) structural aberrations and mutations involving CIITA, the master transcriptional regulator of major histocompatibility complex class II gene transcription (a system that is responsible for antigen presentation7 ) and (2) copy number alterations and rearrangements of the programmed death-ligand 1 (PDL1 and protein CD274) and PDL2 (also known as PDCD1LG2 and protein CD273) genes at 9p24.1 that are responsible for T-cell–mediated immune responses (see figure).8 The Mottok study adds detail to the abnormalities in CIITA and PDL, as well as other immune-related genes, in terms of mutation and copy number information. In addition, mutations in the interferon regulatory factor (IRF) genes, including a hotspot in the B-cell immune response gene IRF4, and mutations in IRF downstream targets were detected in 52% of the cases. Altogether, this multipronged genetic approach, which affects antigen presentation as well as responses from T cells, natural killer cells, and macrophages (see figure) creates an immune-privileged phenotype for PMBL that allows it to fly under the detection radar and represents a blend of various strategies used by other B-cell–derived lymphomas (reviewed in Scott and Gascoyne9 ).
Among driver mutations, Janus kinase-signal transducer and activator of transcription (JAK-STAT) and nuclear factor-κB (NF-κB) signaling pathway genes, including suppressor of cytokine signaling 1 (SOCS1), were most frequently altered in this study. Mottok et al1(Fig1) provide a heat map of driver mutations. Interestingly, the driver gene mutation profile in PMBL was found to be more similar to that in Hodgkin lymphoma than to that in DLBCL, underscoring the previous gene expression observations and potentially blended biology of PMBL, which may represent an intermediate malignancy between Hodgkin and non-Hodgkin lymphoma.
Altogether, Mottok et al provided a detailed characterization of the genomic landscape and correlations with gene expression levels of a large, well-defined cohort of patients with PMBL that should facilitate our understanding of PMBL biology, support the development of accurate diagnostics, and encourage design of therapeutic approaches aimed at restoring tumor immune recognition and targeting other vulnerabilities.9,10
Conflict-of-interest disclosure: L.M.R. is an inventor on a patented, nonlicensed, assay known as the Lymph3Cx.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal