In this issue of Blood, Petit et al demonstrate the clinical utility of a genetic classifier combined with end of induction minimal residual disease (MRD) assessment and presenting white blood cell (WBC) count in the effective risk stratification of pediatric patients with T-cell acute lymphoblastic leukemia (T-ALL).1 The integration of these factors allowed the identification of 3 discrete clinical risk groups providing an opportunity for improved treatment allocation.
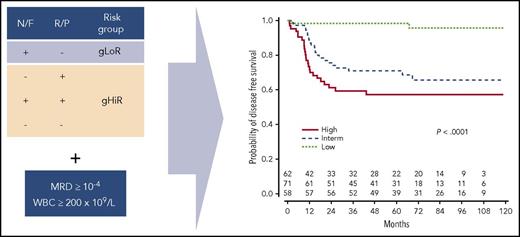
The genomic classifier separates patients into a gLoR or gHiR group based on the presence (+) or absence (−) of Notch1/FBXW7 (N/F) and Ras/PTEN (R/P) mutations. Subsequent integration with MRD status and WBC count allows stratification into 3 discrete clinical risk groups. Interm, intermediate. Adapted from Figure 5 in the article by Petit et al on page 289.
The genomic classifier separates patients into a gLoR or gHiR group based on the presence (+) or absence (−) of Notch1/FBXW7 (N/F) and Ras/PTEN (R/P) mutations. Subsequent integration with MRD status and WBC count allows stratification into 3 discrete clinical risk groups. Interm, intermediate. Adapted from Figure 5 in the article by Petit et al on page 289.
Pediatric ALL has seen a dramatic improvement in outcome over the past 30 years thanks to advances in multiagent chemotherapeutic regimens and risk-defined treatment stratification.2 Most protocols now use some combination of age, WBC, MRD, and genetics to accurately direct treatment intensity. MRD is acknowledged as the single most powerful predictive factor in ALL, successfully guiding both treatment intensification and reduction.3,4 In B-cell ALL, the presence of particular cytogenetic abnormalities, such as ETV6-RUNX1, high hyperdiploidy, and BCR-ABL1, provides further prognostic information. However, despite detailed characterization of recurrent genetic abnormalities,5 T-ALL has so far eluded a similar cytogenetic-based stratification; protocols therefore largely rely on MRD. To address this, the authors performed a detailed retrospective analysis on a cohort of pediatric patients treated for T-ALL on the French Acute Lymphoblastic Leukaemia Study Group 2000 protocol. They used a genetic classification, previously used in adult T-ALL patients,6 which stratifies based on the presence or absence of several recurrent genetic aberrations. Mutations in NOTCH1 and FBXW7 (N/F) both lead to NOTCH1 activation, driving a key oncogenic pathway in T-ALL, and have, in general, been associated with a favorable outcome.7 In contrast, mutations in Ras and alterations in PTEN (R/P) have predominantly been associated with a poorer outcome.8 Combining these mutations in their oncogenetic classifier divided the cohort exactly in half into genetic low-risk (gLoR) and genetic high risk (gHiR) groups. In essence, the gLoR group included only patients with an N/F mutation in the absence of an R/P mutation, whereas all other patients were allocated to the gHiR group. Outcomes were significantly different with a 5-year cumulative incidence of relapse (CIR) of 11.4% (95% confidence interval [CI], 4.8-17.2) in gLoR vs 36.3% (95% CI, 27.2-44.8; P < .0001) in the gHiR group.
To further improve stratification, the authors integrated the genetic classification with MRD and presenting WBC identifying 8 patient subsets, which they simplified into 3 clinical risk groups of roughly equal size, termed low, intermediate, and high based on the risk of relapse (see figure). Most significantly, the low-risk grouping identified one-third of patients with N/F mutations, WBC < 200 × 109/L and good MRD response who had an impressive 5-year CIR of only 1.7% (95% CI, 0-5.1) and who could be considered for treatment reduction. The clinical risk grouping was less successful at clearly separating the remaining patients, with the intermediate- and high-risk groups showing a 5-year CIR of 29.1% (95% CI, 18.3-39.7) and 37.7% (95% CI, 25.3-50.1) respectively. However, it was possible to identify a higher risk subgroup of patients, accounting for 25% of the cohort, with gHiR and high MRD patients who had a relapse risk of >40% irrespective of WBC count.
This study is an important addition to the literature, highlighting the prognostic power of genetics and MRD in pediatric T-ALL. However, there are several limitations that will need to be addressed before the results can be integrated into clinical practice. First, the findings need to be validated in other international cohorts, particularly given previous inconsistency in the prognostic relevance of specific genetic abnormalities in T-ALL, including R/P mutations that did not affect outcome in the UKALL2003 study.9 Second, the use of a later MRD time point, which has been shown to be more discriminative in T-ALL,10 could further refine the risk stratification, particularly in the identification of a true high-risk group. Clearly, selecting this group is only the first step; the real challenge lies in identifying effective treatments for a population that is often refractory to conventional therapy.
Overall, Petit et al provide convincing evidence that, as with B-cell ALL, the future of prognostication in T-ALL lies in the integration of genetic abnormalities and MRD. However, given the relative rarity of T-ALL, which accounts for only 15% of ALL cases in children, future studies will require collaboration across trial groups. The assembly of larger cohorts should permit greater appreciation of the complex interplay of multiple genetic abnormalities and improved discrimination of prognostic subgroups.
Conflict-of-interest disclosure: The author declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal