In this issue of Blood, Dovey et al provide novel insights into the different ways in which the nucleophosmin (Npm1) mutation synergizes with its most common proliferative mutation partners (either Flt3ITD or NrasG12D/+) to give rise to acute myeloid leukemia (AML) with different features in genetically engineered mice.1
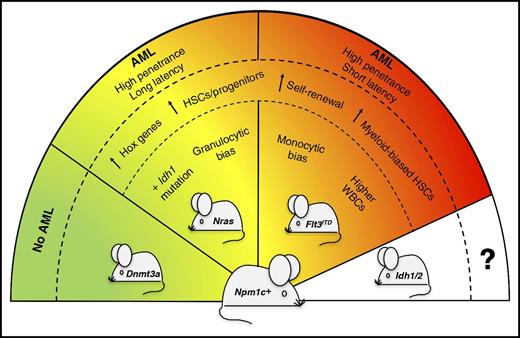
Severity of Npm1cA/+-mutated AML is dictated by concomitant mutations. The presence of different partner mutations in the Npm1cA/+-mutated mouse model (Npm1c+) variably affects the severity and penetrance of the disease phenotype. In particular, Npm1cA/+;NrasG12D/+ mice develop a less aggressive and more differentiated AML than Npm1cA/+;Flt3ITD mice. Compound mutant mice carrying Npm1cA/+;Dnmt3aR878H mutations do not develop leukemia.9 Compound mice co-mutated for Npm1cA/+ and Idh1/2 genes are expected to be generated for future comparative studies. HSCs, hematopoietic stem cells; WBCs, white blood cells.
Severity of Npm1cA/+-mutated AML is dictated by concomitant mutations. The presence of different partner mutations in the Npm1cA/+-mutated mouse model (Npm1c+) variably affects the severity and penetrance of the disease phenotype. In particular, Npm1cA/+;NrasG12D/+ mice develop a less aggressive and more differentiated AML than Npm1cA/+;Flt3ITD mice. Compound mutant mice carrying Npm1cA/+;Dnmt3aR878H mutations do not develop leukemia.9 Compound mice co-mutated for Npm1cA/+ and Idh1/2 genes are expected to be generated for future comparative studies. HSCs, hematopoietic stem cells; WBCs, white blood cells.
NPM1 mutations leading to aberrant cytoplasmic dislocation of nucleophosmin and their frequent co-occurrence with FLT3ITD mutation were first discovered in human AML in a pre–whole-genomic-sequencing era.2 Subsequent studies (reviewed in Falini et al3 ) clearly demonstrated that NPM1 mutations are among the most common genetic lesions in AML (accounting for about one-third of all cases and 50%-60% of AML with normal cytogenetics) and that they are rather specific for AML, mutually exclusive with other genetic lesions defining distinct leukemia entities of the World Health Organization (WHO) classification, and closely associated with unique gene expression profiles and micro-RNA profiles. Moreover, unlike mutations of other genes (eg, DNMT3A), those involving NPM1 are not associated with clonal hematopoiesis, strongly suggesting that they represent gatekeeper mutations critical for establishing the AML phenotype.4 Because of all of these unique features, NPM1-mutated AML has been included as a distinct leukemia entity in the 2016 WHO classification of human myeloid neoplasms.5
The clinical and laboratory features as well as the outcome of the different WHO leukemia entities may be influenced by the presence of 1 or more concomitant mutations; for example, those involving c-Kit that confer a worse prognosis in AML with t(8;21) (q22;q22) RUNX1-RUNX1T1. NPM1-mutated AML is not an exception to this rule, as its association with a high load of FLT3ITD usually results in a bad outcome6 whereas, in patients carrying the NPM1cA/+;NRASG12D/+ double-mutated genotype, a trend toward better survival was reported.7 Similarly, AML patients carrying mutations of all 3 NPM1, DNMT3A, and FLT3 genes display a much worse prognosis than those with the triple-mutated genotype NPM1cA/+;DNMT3A+;NRASG12D/+.8
The study by Dovey et al in genetically engineered mice sheds further light on the cellular and molecular mechanisms underlying these clinical observations in AML patients (see figure). When comparing the short-term impact of double mutations (Npm1cA/+;Flt3ITD vs Npm1cA/+;NrasG12D/+) on hematopoiesis, the 2 mouse models shared not only similarities but also important differences. Features that were common to both the Npm1cA/+;Flt3ITD and Npm1cA/+;NrasG12D/+ compound mice included the overexpression of Hox genes and the increase of myeloid-biased hematopoietic stem/progenitor cells with a higher self-renewal capacity. On the other hand, as compared with the Npm1cA/+;NrasG12D/+ compound model, the Npm1cA/+;Flt3ITD mice showed higher peripheral blood white cell counts due to increased monocyte growth (different from the granulocytic bias of Npm1cA/+;NrasG12D/+ mutants) and early decrease of common lymphoid progenitors.
Monitoring aged animals, the authors demonstrated for the first time that Npm1cA/+;NrasG12D/+ mutations can cooperate to drive highly penetrant AML in mice, although the disease was less aggressive and more differentiated than that occurring in Npm1cA/+;Flt3ITD mice. This is in contrast with the lack of frank AML development in the recently reported Npm1cA/+;Dnmt3aR878H compound model,9 suggesting that the function of the co-mutated gene dictates whether leukemogenesis occurs or not.
Using CRISPR-Cas9 to disrupt selected genes in Cas9-expressing primary mouse AML cells, the authors provided functional evidence of the critical role played by HoxA genes for the survival of both compound mice models, thus confirming previous reports on the importance of HOX gene overexpression in NPM1-driven human AML.3,10 In both models, AML evolution was associated with the acquisition of additional copies of the mutant FLT3ITD or NrasG12D/+ alleles, through copy-neutral loss-of-heterozygosity (LOH) or copy number gain. However, new somatic mutations in AML driver genes (eg, Idh1 and ptpn11) were detected only in the Npm1cA/+;NrasG12D/+ compound mice.
Collectively, these experimental findings provide an explanation for the higher frequency, aggressiveness, and worse prognosis of the NPM1cA/+;FLT3ITD vs NPM1cA/+;NRASG12D/+ genotype in AML patients, and further support the WHO concept of NPM1-mutated AML being a distinct leukemia entity5 whose clinicopathological findings and outcome may vary according to the accompanying genetic lesion(s). This raises the question of why, unlike that observed in mice, the copy-neutral LOH is much more common in FLT3ITD- than in NRAS-mutated human AML. Possible explanations may be the differences in human vs mouse synteny and/or the fact that mitotic recombination events leading to LOH are more likely to occur in inbred mice.
Which are the mechanistic perspectives and therapeutic applications for these innovative mouse models of NPM1-mutated AML? They represent a relevant platform for investigating the nature of specific preleukemic populations that cannot be found in humans because, in this clinical setting, NPM1/FLT3 and NPM1/NRAS co-mutations only present as frank AML. These models can also be used to further improve treatment strategies in AML, by assessing the efficacy of novel antileukemic agents used alone or in combination and studying the mechanisms of resistance. Finally, the generation of as-yet-unavailable compound mice co-mutated for NPM1 and IDH1/2 genes is also expected to be an invaluable research tool.
Conflict-of-interest disclosure: B.F. applied for a patent on the clinical use of NPM1 mutants. P.S. declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal