To the editor:
Monocytes originate from the bone marrow (BM), are distributed in the bloodstream, and can differentiate in the tissue into skin macrophages or intestinal dendritic cells (DCs).1 They play an essential role in the defense against pathogens2 and are implicated in a range of diseases.3
Labeling experiments with 6,6-2H2-glucose4 and 3H-thymidine5,6 have suggested a monocyte residence time in blood of 2 to 4 days. Although these experiments considered monocytes as a single population, the currently held view is that at least 3 monocyte subsets exist: classical CD14++CD16– monocytes (CMs), intermediate CD14++CD16+ monocytes (IMs), and nonclassical CD14+CD16++ monocytes (NCMs). The residence times of these subsets in blood and their interrelationship remain to be determined. The 3 subsets are characterized by the gradually changing expression of surface markers,7 differential gene enhancer profiles8 expression profiles,9,10 and differences in functionality.3,11 Their maturation kinetics also differ, because human CMs repopulate the bloodstream first after hematopoietic stem cell transplantation, followed by IMs and later by NCMs.12 In rhesus macaques, similar results were found by using in vivo bromodeoxyuridine labeling.13 The gradually changing expression patterns, combined with their consecutive repopulation/labeling kinetics has led to the prevailing idea that monocytes differentiate from CMs via IMs to NCMs.9,13
In mice, there is more direct evidence for such a linear differentiation pattern.14 Adoptively transferred CM homologs were shown to differentiate into NCMs,15,16 and in vivo imaged monocytes were shown to lose CM marker CCR2 while acquiring NCM marker CXC3CR1 at sites of sterile inflammation.17 However, adoptively transferred CM preparations might contain small numbers of monocyte progenitors15 and thus, even in mice, there remains discussion on the developmental relation of these monocyte subsets under homeostatic conditions.3,15,18
To investigate the circulatory and maturation/differentiation kinetics of the human monocyte subsets, we used in vivo 6,6-2H2-glucose labeling in 14 volunteers19,20 (see supplemental Methods, available on the Blood Web site). This study was performed after receiving approval from the local ethics review board (Medisch Ethische Toetsingscommissie Utrecht) and obtaining written informed consent from all volunteers in accordance with the Declaration of Helsinki. Our study population consisted of 5 healthy volunteers and 9 eosinophilic asthma patients. Because no differences in monocyte subset counts or labeling kinetics were found between asthma patients and healthy volunteers (supplemental Figure 1), we combined data from both groups for analysis.
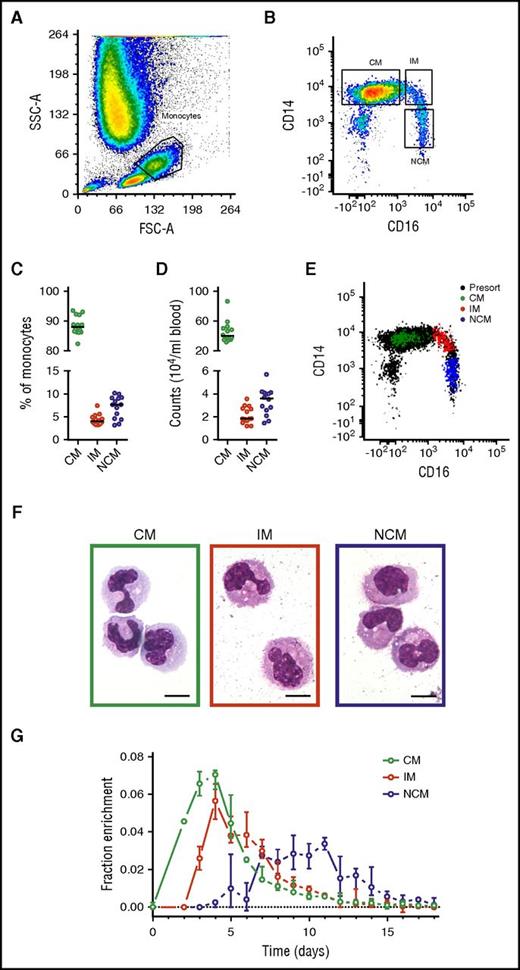
Monocyte subset numbers and percentages in blood (Figure 1) were in agreement with previous findings,3 with a median of 89% CMs, 4% IMs, and 7% NCMs. DNA 2H-enrichment was first detected in CMs, in which it peaked at days 3 to 4, similar to earlier labeling studies on the whole monocyte compartment.4,5 In IMs and NCMs, label enrichment was detected later and peaked after approximately 4 and 8 days, respectively. These findings are in line with previous results suggesting a linear differentiation pattern.12,13
Analysis and isolation of circulating monocytes. Monocyte subsets were gated on the basis of (A) forward scatter (FSC) and side scatter (SSC) and (B) singlets and CD14/CD16 expression. Resulting monocyte (C) percentages and (D) numbers showed CMs to be the most common circulating monocyte subset, followed by NCMs and IMs. (E) Fluorescence-activated cell sorter analysis of 200 sorted cells from all 3 monocyte subsets typically revealed a purity >99% and good separation between the populations. Note that the inclusion of pan-monocyte markers CD86 and major histocompatibility complex II in our gating strategy (either with or without the inclusion of cells in the lymphocyte gate) resulted in a slightly higher number of CMs and IMs recovered (1% and 2% higher counts on average, respectively) and a lower number of recovered NCMs (3%). However, use of these cell numbers did not result in significant differences in parameter estimates. (F) Cytospin preparations stained with May-Grünwald-Giemsa showed an increasingly mature phenotype from CMs to NCMs as characterized by a more neutrophilic cytoplasm and increasingly dendritic appearance. The objective was a 100× oil immersion lens, numerical aperture was 1.30, and scale bars = 10 μm. (G) DNA 2H-enrichments after in vivo pulse-labeling with 6,6-2H2-glucose were determined by gas chromatography-mass spectrometry and plotted after normalization for plasma enrichment and intracellular dilution of label (supplemental Methods). Results are based on a total of 249 measurements from 14 individuals; circles indicate medians, and bars represent interquartile ranges. (A-B,E-F) Representative results from 1 experiment. (C-D) Circles represent the medians of 6 measurements for each volunteer, and lines indicate medians for the 14 volunteers.
Analysis and isolation of circulating monocytes. Monocyte subsets were gated on the basis of (A) forward scatter (FSC) and side scatter (SSC) and (B) singlets and CD14/CD16 expression. Resulting monocyte (C) percentages and (D) numbers showed CMs to be the most common circulating monocyte subset, followed by NCMs and IMs. (E) Fluorescence-activated cell sorter analysis of 200 sorted cells from all 3 monocyte subsets typically revealed a purity >99% and good separation between the populations. Note that the inclusion of pan-monocyte markers CD86 and major histocompatibility complex II in our gating strategy (either with or without the inclusion of cells in the lymphocyte gate) resulted in a slightly higher number of CMs and IMs recovered (1% and 2% higher counts on average, respectively) and a lower number of recovered NCMs (3%). However, use of these cell numbers did not result in significant differences in parameter estimates. (F) Cytospin preparations stained with May-Grünwald-Giemsa showed an increasingly mature phenotype from CMs to NCMs as characterized by a more neutrophilic cytoplasm and increasingly dendritic appearance. The objective was a 100× oil immersion lens, numerical aperture was 1.30, and scale bars = 10 μm. (G) DNA 2H-enrichments after in vivo pulse-labeling with 6,6-2H2-glucose were determined by gas chromatography-mass spectrometry and plotted after normalization for plasma enrichment and intracellular dilution of label (supplemental Methods). Results are based on a total of 249 measurements from 14 individuals; circles indicate medians, and bars represent interquartile ranges. (A-B,E-F) Representative results from 1 experiment. (C-D) Circles represent the medians of 6 measurements for each volunteer, and lines indicate medians for the 14 volunteers.
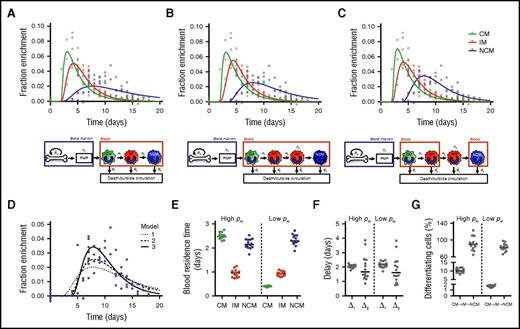
Mathematical models (supplemental Methods) were fitted to the measured 2H-enrichment levels to estimate monocyte subset kinetics. These models take into account the observed ratios between cell numbers in each subset (assumed to be constant) and assume that incorporation of label occurred only at the monocyte progenitor stage in the BM. After maturation in the postmitotic pool (PMP), the models assume that cells enter the bloodstream as CMs and subsequently mature into IMs and NCMs. Assuming differentiation from IMs into NCMs directly in the circulation gave reasonable fits to the enrichment data of CMs and IMs but failed to describe the delay with which label was observed in NCMs (Figure 2A). We therefore extended the model by assuming that IMs can mature into NCMs only after a delay of Δ2 days, Assuming that this maturation step occurs inside the blood gave an improved fit to the data (Figure 2B; moderate improvement according to the Kullback-Leibler’s scale; supplemental Methods), but an even better fit was obtained when we assumed that this differentiation step occurs outside the blood (Figure 2C).
Linear differentiation models used to describe the data. Three different models were fitted to the (nonnormalized) DNA 2H-enrichment data. Because fits with a high or low rate of cell division in the BM (pm) are very similar, only the predicted curves with an estimated high pm are shown. The first model (A) assumes simple linear differentiation of CMs to IMs to NCMs in the blood. The other 2 models assume that IMs can mature into NCMs only after a delay of Δ2 days. The second model (B) assumes that this maturation step occurs in the blood, whereas in the third model (C), this takes place outside the circulation. Because the largest differences between the models lay in the fits of the NCMs, predicted curves for NCMs were plotted separately in panel D to allow for easier comparison. The third model produced the best fits (lowest Akaike information criterions), and thus it was used to estimate (E) the residence times of monocytes in the different monocyte subsets, (F) the delays in the PMP and between IM and NCM differentiation, and (G) the percentages of cells that differentiate from CMs into IMs or from IMs into NCMs (supplemental Methods). Data were obtained from 14 individuals, and both data and curves were normalized for plasma enrichment and intracellular dilution of label (supplemental Methods). Individual fits are shown in supplemental Figure 3 and the corresponding parameter estimates in supplemental Tables 1, 2, and 3.
Linear differentiation models used to describe the data. Three different models were fitted to the (nonnormalized) DNA 2H-enrichment data. Because fits with a high or low rate of cell division in the BM (pm) are very similar, only the predicted curves with an estimated high pm are shown. The first model (A) assumes simple linear differentiation of CMs to IMs to NCMs in the blood. The other 2 models assume that IMs can mature into NCMs only after a delay of Δ2 days. The second model (B) assumes that this maturation step occurs in the blood, whereas in the third model (C), this takes place outside the circulation. Because the largest differences between the models lay in the fits of the NCMs, predicted curves for NCMs were plotted separately in panel D to allow for easier comparison. The third model produced the best fits (lowest Akaike information criterions), and thus it was used to estimate (E) the residence times of monocytes in the different monocyte subsets, (F) the delays in the PMP and between IM and NCM differentiation, and (G) the percentages of cells that differentiate from CMs into IMs or from IMs into NCMs (supplemental Methods). Data were obtained from 14 individuals, and both data and curves were normalized for plasma enrichment and intracellular dilution of label (supplemental Methods). Individual fits are shown in supplemental Figure 3 and the corresponding parameter estimates in supplemental Tables 1, 2, and 3.
For all models, we found 2 optima, 1 with slow and 1 with fast dynamics of BM precursors. The estimated average maturation time in the PMP (Δ1) was consistently ∼2 days. For the third best-fitting model, monocytes were estimated to reside in the blood as CMs for 0.4 or 2.5 days, depending on the specific optimum, with corresponding turnover rates of monocyte precursors in the BM of 0.4 or 2.6 per day, respectively. The former optimum with a relatively slow BM turnover gave a small improvement of the fit to the data compared with the latter (supplemental Table 1). To determine which of the optima is the one that matches the biology, information about the dynamics of monocyte progenitors or the relative pool sizes of progenitors and circulating monocytes21 would be required. According to the literature,22 the number of circulating monocytes exceeds by far the number of progenitors, which would suggest that monocyte progenitors have a fast turnover rate and that CMs have a residence time of 2.5 days in the blood. The residence times of IMs and NCMs could be estimated with more certainty, because both optima yielded similar estimates of ∼0.9 days for IMs and ∼2.3 days for NCMs. Likewise, we consistently estimated IMs to stay outside the circulation for ∼1.6 days with negligible cell death before reentering the blood as NCMs.
In addition to estimating monocyte subset kinetics, our model allowed estimation of the fraction of cells lost from circulation during each maturation step. We estimated that, on average, less than 10% of all CMs differentiate into circulating IMs, whereas 82% to 89% of circulating IMs eventually mature into circulating NCMs. This is in line with previous experiments showing monocyte-derived macrophages in skin23 and DCs in the gut16 to be directly derived from CMs, implying that not all CMs become IMs. Alternatively, there might be noncirculating pools of IMs and NCMs, which cannot be obtained by venipuncture. The existence of noncirculating monocytes has been demonstrated in the BM24 in the form of patrolling NCMs11 adhering to blood vessel walls or as marginated monocytes in mice. 2H-labeling experiments that include noncirculating monocyte samples might allow determination of the location of IM into NCM maturation.
Note that our models assume the existence of no more than 2 kinetically different populations within each subset. Additional monocyte subsets as proposed recently25 might result in slightly different estimates. Furthermore, the fact that our results can be fitted with a linear CM-IM-NCM differentiation does not exclude the possibility that each subset develops separately in the BM. This would require longer retention of IMs and NCMs in the PMP and can be excluded only by lineage tracing experiments (eg, by infusion of labeled CMs followed by measurements in IMs and NCMs).
Our results have important implications for monocytes under homeostasis. We show that all 3 monocyte subsets have a rapid turnover in the circulation. Our results support the hypothesis of a linear CM-IM-NCM differentiation but imply that the majority of CMs do not end up as circulatory NCMs and suggest that the last differentiation step takes place outside the circulation. Thus, monocytes seem to leave the circulation as IMs and re-enter as NCMs. From these results, important questions arise, such as what determines whether a monocyte follows the CM-IM-NCM differentiation pathway, why IMs leave the circulation, and how these processes are involved in immune homeostasis and pathogenesis of immune-mediated diseases.
The online version of this article contains a data supplement.
Authorship
Acknowledgments: The authors thank Simone Sluis-Eising for taking care of our healthy volunteers and patients, Corneli van Aalst for help in the laboratory, Sigrid Otto for her help with the gas chromatography–mass spectrometry analyses, the fluorescence-activated cell sorter operators of the Laboratory for Translational Immunology Flow Cytometry Core Facility, and Stijn van Oirschot and Marieke Nühn for their measurements for the revision of the manuscript.
This work was supported by grant 3.2.10.052 from the Dutch Lung Foundation.
Contribution: T.T. and L.C. included volunteers and isolated cells; T.T. performed all other experimental procedures and analyses; J.D. wrote the mathematical model with help from J.A.M.B. and R.J.d.B.; T.T. and J.D. fitted the models to the data; T.T. wrote the first draft of the manuscript, which was revised with help of L.K., K.T., and all other authors; and all authors contributed to the design, interpretation, and coordination of the study and read and approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for T.T. is Quantitative Immuno-Hematology, Institut Curie, PSL Research University, CNRS UMR168, Paris, France.
The current affiliation for L.C. is Department of Pulmonary Diseases, Maastricht UMC, Maastricht, The Netherlands.
Correspondence: Kiki Tesselaar, Department of Immunology, Laboratory for Translational Immunology, University Medical Centre Utrecht, KC 02.085.2, P.O. Box 85090, 3508AB Utrecht, The Netherlands; e-mail: k.tesselaar@umcutrecht.nl.
References
Author notes
J.A.M.B. and K.T. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal