Key Points
Pu.1 and Spi-b form vertical and paralleled regulatory networks to orchestrate RBI- and VDA-born macrophage development, respectively.
The vertical and paralleled Pu.1−Spi-b regulatory networks control the development of RBI- and VDA-born macrophages by regulating Irf8.
Abstract
Macrophages are key components of the innate immune system and play pivotal roles in immune response, organ development, and tissue homeostasis. Studies in mice and zebrafish have shown that tissue-resident macrophages derived from different hematopoietic origins manifest distinct developmental kinetics and colonization potential, yet the genetic programs controlling the development of macrophages of different origins remain incompletely defined. In this study, we use zebrafish, where tissue-resident macrophages arise from the rostral blood island (RBI) and ventral wall of dorsal aorta (VDA), the zebrafish hematopoietic tissue equivalents to the mouse yolk sac and aorta-gonad-mesonephros for myelopoiesis, to address this issue. We show that RBI- and VDA-born macrophages are orchestrated by distinctive regulatory networks formed by the E-twenty-six (Ets) transcription factors Pu.1 and Spi-b, the zebrafish ortholog of mouse spleen focus forming virus proviral integration oncogene B (SPI-B), and the helix-turn-helix DNA-binding domain containing protein Irf8. Epistatic studies document that during RBI macrophage development, Pu.1 acts upstream of Spi-b, which, upon induction by Pu.1, partially compensates the function of Pu.1. In contrast, Pu.1 and Spi-b act in parallel and cooperatively to regulate the development of VDA-derived macrophages. Interestingly, these two distinct regulatory networks orchestrate the RBI- and VDA-born macrophage development largely by regulating a common downstream gene, Irf8. Our study indicates that macrophages derived from different origins are governed by distinct genetic networks formed by the same repertoire of myeloid-specific transcription factors.
Introduction
Macrophages are a subclass of innate immune cells that reside in almost all tissues of vertebrates.1-4 The importance of this population of cells is exemplified by their broad involvement in organogenesis, tissue regeneration and remodeling, and immune defense, and perturbation of their development and functions often leads to detrimental consequences, resulting in occurrence of various disorders such as leukemia, infectious diseases, and autoimmune diseases. Therefore, a comprehensive understanding of how tissue-resident macrophages are formed and how they function will be clinically beneficial to cure macrophage-associated disorders.
In mammals, macrophages are predominantly derived from the hematopoietic precursors born in the yolk sac (YS) and the aorta-gonad-mesonephros (AGM), where embryonic and adult hematopoiesis occurs.5-9 Recent cell fate–mapping studies with the conventional promoter-directed CreER-loxP system have indicated that macrophages derived from the YS and AGM manifest distinct colonization potential and tissue preferences.10-12 Likewise, macrophages originated from the rostral blood island (RBI) and ventral wall of dorsal aorta (VDA), the zebrafish hematopoietic tissue equivalent to the mammalian YS and AGM for myelopoiesis, respectively,13-16 colonize the central nervous system with distinct colonization kinetics and potential.17 However, the genetic programs governing the development of these different macrophage populations and their cellular functions remain unclear.
It is well known that many key steps of macrophage development are dictated by tissue-specific transcription factors.18 PU.1, the founding member of the spleen focus forming virus proviral integration oncogene (SPI) subfamily of E-twenty-six (ETS) transcription factors, is one of the best-characterized tissue-specific transcription factors involved in macrophage development.19,20 PU.1-null mice die either in late gestation or shortly after birth and display profound defects in myeloid cell formation during early embryogenesis.19,20 Subsequent studies in mice and zebrafish have revealed that PU.1 regulates specification and maturation of macrophages and neutrophils in a dosage-dependent manner.21,22 Because PU.1-null mice cannot survive to adulthood and spatial-specific lineage tracing in PU.1 knockout mice is still lacking, it remains unclear whether PU.1 is differentially required for different waves of macrophage development. In addition to PU.1, the mammalian SPI subfamily of ETS transcription factors includes 2 other members, SPI-B and SPI-C. Interestingly, although SPI-B shares the highest degree of similarity with PU.1, knockout studies in mice have shown that SPI-B is dispensable for the development of the macrophage lineage.23 By contrast, SPI-C is required for the development of red pulp macrophage in spleen.24 Similar to mammals, 3 members of the Spi subfamily (Pu.1, Spi1-like and Spi-c) have been annotated in zebrafish. Genetic ablation and morpholino (MO) knockdown studies have shown that Pu.1 is the functional ortholog of mammalian PU.1,21,25 whereas Spi1-like has been shown to play a role in embryonic myelopoiesis.26 Intriguingly, our recent study showed that while RBI-born (embryonic) macrophages are depleted in hypomorphic pu.1G242D mutant zebrafish, VDA-derived (adult) macrophages remain largely unaffected,17 suggesting that the development of RBI- and VDA-born macrophages is differentially regulated by Pu.1 and implying that additional factors are involved in the regulation of VDA-born macrophages.
In this study, we used reverse genetics and fate-mapping analysis to dissect the genetic programs governing the development of RBI- and VDA-born macrophages. We showed that while pu.1-null mutation is sufficient to block the formation of RBI-derived macrophages, simultaneous inactivation of Pu.1 and Spi-b is necessary to abolish the development of VDA-derived macrophages. Epistatic analysis revealed that Pu.1 and Spi-b act in parallel and cooperatively to regulate VDA-derived macrophage development, whereas Pu.1 acts upstream of Spi-b during the development of RBI-derived macrophages. We further documented that Irf8 is a common key downstream target of these distinct Pu.1−Spi-b genetic loops for both RBI- and VDA-derived macrophage development.
Methods
Zebrafish
Zebrafish were maintained according to standard protocol.27 AB wild-type, pu.1G242D,21 pu.1Δ839, pu.1Δ371, spi-bΔ232, irf8Δ171, Tg(zpu.1:eGFP)df5,28 Tg(hsp70:mCherry-T2aCreERT2)#12,29 Tg(coro1a:eGFP)hkz05t,30 Tg(mpeg1:loxP-DsRedx-loxP-eGFP)hkz015t,31 and Tg(mpeg1:loxP-eGFP) lines were used in this study.
Reverse gene targeting with TALENs
Transcription activator-like effector nuclease (TALEN) constructs were generated using the “unit assembly” method.32 Synthesis of capped TALEN messenger RNA (mRNA) was performed with the Sp6 mMESSAGE mMACHINE Kit (Ambion) according to the manufacturer’s instructions.
Mutant genotyping
pu.1G242D genotyping was performed by BsmFI digestion as described previously.17 The other mutants (pu.1Δ839, pu.1Δ371, spi-bΔ232, and irf8Δ171) were identified by DNA electrophoresis of polymerase chain reaction (PCR)–amplified genomic DNA. The following primers were used for PCR amplification: pu.1Δ839 or pu.1Δ371, 5′-CCTTGTGTAGACTTCGATGAACTG-3′/5′-CGTTCGCTTGTGTGTTAGTGTCAC-3′; Spi-bΔ232, 5′-GGTCATGGCCAAGACTTGATC-3′/5′-CTAAACTG TGCAGGGCTTCGG-3′; and irf8Δ171, 5′-CTGCTTGGATGCCGTGAGTATG-3′/5′-CAT GTACACAGGGATCTATGATC-3′.
Region-specific cell labeling with the IR-LEGO-CreER-loxP system
IR-LEGO-CreER-loxP labeling was performed as previously described.17 In brief, embryos were mounted in methylcellulose and subjected to heat shock with a 1345-nm infrared laser in the RBI and VDA, respectively. The power of the 1340-nm laser was ∼180 to 200 mW, and the duration of each heat shock was 30 seconds. Three spots in one side of the RBI of 12– to 18–hours postfertilization (hpf) embryos and 4 spots in the VDA of 28- to 30-hpf embryos were irradiated. Immediately after the irradiation, the embryos were treated with 4-OHT for 17 to 22 hours, and the labeled cells were marked by GFP expression.
Generation of transgenic lines
The 9-kb promoter of pu.128 and flag-myc double-tagged zebrafish pu.1 or spi-b were cloned into the modified pBluescript II SK(+) vector containing 2 arms of Tol2 sequence. The resulting -9kbpu.1:flag-myc-pu.1 or -9kbpu.1:flag-myc-Spi-b construct, together with transposase mRNA,33 was injected into 1-cell-stage embryos. The injected embryos were raised to adulthood for further screening of germline transmission.
In vitro synthesis of antisense RNA probes and WISH
Antisense RNA probes and whole-mount in situ hybridization (WISH) were performed according to standard protocol.27
Histology
Neutral red staining
Neutral red staining was performed as previously described.14
Acridine orange and TUNEL staining
Acridine orange and terminal deoxynucleotidyltransferase-mediated dUTP nice end labeling (TUNEL) staining was performed according to manufacturer’s instruction or as previously reported.35
Cryosection
All the samples were fixed with 4% paraformaldehyde for 1 or 2 days and washed with PBS for another day. For 15- and 30–days postfertilization (30-dpf) juveniles, the fixed fish were directly subjected to cryosection at 30-µm intervals and subsequently costained with GFP/DsRed and Lcp1 antibodies. For adult fish, the brains of the fixed fish were dissected for cryosection and antibody staining.
Imaging
Fluorescent signals from live reporter lines, FISH, or immunostaining were imaged under a Leica SP8 confocal microscope. The HC PL APO 20×/0.70 DRY objective was used in this study.
Cell dissociation for FACS
To dissociate cells from zebrafish embryos or juveniles, 22G or 24G needles were used to break up the tissues, followed by 37°C digestion with dispase (5% final, Sigma) for 20 min. The dissociated cells were re-suspended with 400 to 600 µL cold Hank’s balanced salt solution, passed through 40-µm nylon filters, and finally subjected to fluorescence-activated cell sorting (FACS) with a Becton Dickinson FACSAria IIIu.
RNA and cDNA preparation
Total RNA were extracted from FACS-sorted cells with NucleoSpin RNA XS kit (Macherey-Nagel) and subjected to reverse transcription (RT) and amplification with the REPLI-g WTA Single Cell Kit (QIAGEN) according to the manufacturer’s instructions. For large amount of samples, total RNA was extracted with TRIzol reagent or the RNeasy Mini Kit (QIAGEN), and complementary (cDNA) was synthesized with SuperScript III reverse transcriptase (Thermo Fisher).
Quantitative PCR
Quantitative PCR was performed with SYBR Green Supermix (Roche) on a 7500 Fast Real-time PCR system (ABI) and analyzed using the ΔΔCt method. The following primers were used for quantitative PCR: elf1a, 5′-CTTCTCAGGCTGACTGTGC-3′/5′-CCGCTAGCATTACCCTCC-3′; lyz, 5-GCCTGTTCAGACTTGCTTAACG-3′/5′-CAGGCTCGGAGGCTTTGTTTG-3′; mpo, 5′-GCTATACCAGGTTATAATGCATG-3′/5′-CCACAACCTATCGCCATCTCG-3′; mfap4, 5′-GGATGGACGGTGATTCAGAG-3′/5′-CAGATAAAGAGTCGCCTGCT-3′; mpeg1, 5′-CTCCACAGAAAACCAGCGCA-3′/5′-CGTCAGCGATTTCTTCTGCC-3′; pu.1, 5′-GACAGTCAGAACGATCACTCTT-3′/5′-GGAGAGGAGATGGCTGGACG-3′; Spi-b, 5′-GACTCATCCAGAGAGCGGATG-3′/5′-ACAGCGGATGGGAGATGTAAG-3′; and irf8, 5′-CTGCTTGGATGCCGTGAGTATG-3′/5′-ACCTAACTTTTGTTCCTCCTCTG-3′.
Western blotting
Myc-tagged fusion mRNA for each gene were coinjected with eGFP mRNA into 1-cell-stage zebrafish embryos. At 6 hpf, ∼100 embryos were washed with 500 µL PBS and passed through a 100-µL pipette to remove the YS. After centrifugation at 200g for 5 minutes, the pellets were resuspended with 100 µL SDS lysis buffer (63 mM Tris-HCl, pH 6.8, 10% glycerol, 5% β-mercaptoethanol, and 0.002% bromophenol blue). After centrifugation at 13 000 rpm for 10 minutes, 5 µL supernatant was loaded for immunoblotting.36 Anti-Myc antibody (Santa Cruz Biotechnology) and anti-GFP antibody (a kind gift from Robert Qi in the Hong Kong University of Science and Technology) were used as primary antibodies.
Statistics
Statistics were analyzed using the 2-tailed Student t test. A result was considered significant if P < .05. Values represent mean ± standard error of the mean.
Results
Inactivation of Pu.1 blocks RBI-, but not VDA-derived, macrophage development
Previous studies have revealed that reducing Pu.1 activity in hypomorphic pu.1G242D mutant zebrafish blocks the development of RBI-derived macrophages but has little effect on the generation of VDA-derived macrophages.17 This observation suggests that perhaps Pu.1 is dispensable for the VDA-derived macrophage development. To confirm this notion, we generated an amorphic pu.1 allele pu.1∆839 (supplemental Figure 1A, available on the Blood Web site), in which an 839-bp genomic fragment of the pu.1 gene was deleted, leading to a 194-bp deletion in exon 4 (supplemental Figure 1B) and synthesis of an unstable truncated Pu.1∆839 protein lacking the Q-rich transcriptional activation, PEST, and Ets DNA-binding domains (supplemental Figure 1C-F). Thus, pu.1∆839 likely represents a null allele. Phenotypic characterization revealed loss of macrophages in pu.1∆839 mutants during early development as indicated by the lack of mfap4 staining in the periphery at 30 hpf and the loss of neutral red staining in the brain at 3 dpf (Figure 1A-D). In addition, neutrophils (lyz+ cells) and early myeloid progenitors (cebpα+ cells) were also dramatically reduced in pu.1∆839 mutants (Figure 1E-H). Since these early macrophages and neutrophils are derived from the RBI,13,14,16,37 these data suggest that loss of Pu.1 blocks the development of RBI-born macrophages and neutrophils. Consistent with the null mutation, the phenotypes in pu.1∆839 mutants are significantly more severe than the myeloid defects observed in a hypomorphic pu.1G242D mutant, where neutrophils and early myeloid progenitors are intact.21 However, macrophages (as indicated by macrophages on the skin and microglia in the brain) in pu.1∆839 mutants recovered from 5 dpf onwards (supplemental Figure 2A-D). The recovered cells were likely to be able to differentiate into mature macrophages as they displayed typical branched morphology (supplemental Figure 2A′-B′) and could phagocytose bacteria (supplemental Figure 2E-F). Likewise, neutrophils also became evident from 4 dpf onwards (Figure 1L-M). The recovery of macrophages in pu.1∆839 mutants is similar to that observed in hypomorphic pu.1G242D mutants,17 suggesting that complete loss of Pu.1 function fails to block VDA-born macrophage development. To prove this is indeed the case, we outcrossed pu.1∆839 with Tg(hsp70:mCherry-T2a-CreERT2;mpeg1:loxP-DsRedx-loxP-eGFP) (we herein refer to loxP-DsRedx-loxP-eGFP as LRLG) reporter line, in which CreER recombinase is induced by spatial-restricted infrared light–mediated heat shock and subsequently activated by tamoxifen, resulting in the conversion of mpeg1-DsRed into mpeg1-GFP.17 The Tg(hsp70:mCherry-T2a-CreERT2;mpeg1:LRLG);pu.1∆839 mutants were then used to determine the source of the recovered macrophages in pu.1∆839 mutants using infrared light–induced IR-LEGO-CreER-loxP system.17 Results showed that the recovered macrophages (GFP+ cells) in pu.1∆839 mutants were exclusively derived from the VDA (Figure 1I-K). From these observations, we conclude that Pu.1 is essential for the development of RBI-derived macrophages but is dispensable for those derived from the VDA.
Pu.1 is required for the RBI- but not VDA-derived macrophage development. (A-B) WISH of mfap4 expression in 30-hpf siblings (n = 31/31) and pu.1∆839 mutants (n = 12/12). (C-D) Neutral red staining of microglia in 3-dpf siblings and pu.1∆839 mutants. (E-F) WISH of lyz expression in 30-hpf siblings (n = 35/35) and pu.1∆839 mutants (n = 13/13). (G-H) WISH of cebpa expression in 20-hpf siblings (n = 42/42) and pu.1∆839 mutants (n = 13/13). (I-J) Fluorescence imaging of RBI- and VDA-labeled pu.1∆839;Tg(mpeg1:Loxp-Dsred-loxp-GFP) embryos at 8 dpf. Embryos are laterally orientated to view macrophages in the trunk region. (K) Quantification of RBI- or VDA-labeled macrophages (GFP+) in the total recovered macrophages (mpeg1+) in 8 dpf pu.1∆839 mutants compared with control embryos without heat shock. ***P < .001 (Student t test, GFP+/mpeg1+control (mean/standard error [SE]/n) = 0.09/0.01/9), GFP+/mpeg1+RBI (mean/SE/n) = 0.08/0.02/4, GFP+/mpeg1+VDA (mean/SE/n) = 0.66/0.04/7). Error bars represent SE. (L-M) Sudan black B (SB) staining of neutrophils in 4-dpf siblings and pu.1∆839 mutants.
Pu.1 is required for the RBI- but not VDA-derived macrophage development. (A-B) WISH of mfap4 expression in 30-hpf siblings (n = 31/31) and pu.1∆839 mutants (n = 12/12). (C-D) Neutral red staining of microglia in 3-dpf siblings and pu.1∆839 mutants. (E-F) WISH of lyz expression in 30-hpf siblings (n = 35/35) and pu.1∆839 mutants (n = 13/13). (G-H) WISH of cebpa expression in 20-hpf siblings (n = 42/42) and pu.1∆839 mutants (n = 13/13). (I-J) Fluorescence imaging of RBI- and VDA-labeled pu.1∆839;Tg(mpeg1:Loxp-Dsred-loxp-GFP) embryos at 8 dpf. Embryos are laterally orientated to view macrophages in the trunk region. (K) Quantification of RBI- or VDA-labeled macrophages (GFP+) in the total recovered macrophages (mpeg1+) in 8 dpf pu.1∆839 mutants compared with control embryos without heat shock. ***P < .001 (Student t test, GFP+/mpeg1+control (mean/standard error [SE]/n) = 0.09/0.01/9), GFP+/mpeg1+RBI (mean/SE/n) = 0.08/0.02/4, GFP+/mpeg1+VDA (mean/SE/n) = 0.66/0.04/7). Error bars represent SE. (L-M) Sudan black B (SB) staining of neutrophils in 4-dpf siblings and pu.1∆839 mutants.
Inactivation of Pu.1 and Spi-b severely impairs the development of VDA-born macrophages
The normal development of VDA-born macrophages and neutrophils in pu.1∆839 mutants suggests that additional factors, perhaps other Ets transcription members, may compensate for the loss of Pu.1 function in the development of VDA-derived myelopoiesis. We therefore turned our attention to Spi1-like, which is thought to be a co-ortholog of mammalian PU.1 and has been shown to be involved in early myeloid development in zebrafish.26 Intriguingly, our phylogenic tree analysis and protein sequence alignment showed that Spi1-like shares the highest similarity with the mammalian SPI-B among Ets family transcription factors in zebrafish (Figure 2A-B). Furthermore, critical amino acids in the acidic subdomain and Q-rich region of the PU.1 transactivation domain38 were absent in Spi1-like protein (Figure 2C, underlined black letters). Instead, a unique P/S/T-rich region known to be essential for mammalian SPI-B transactivation activity39 is present in Spi1-like (Figure 2C, dashed black box). Thus, Spi1-like is likely to represent the zebrafish ortholog of mammalian SPI-B and is herein designated as Spi-b. To dissect the role of Spi-b, we generated a null allele spi-b∆232, which carried a 232-bp deletion and a 4-bp insertion in exon 5, resulting in the production of a truncated Spi-b∆232 protein lacking 78 amino acids in the Ets domain (supplemental Figure 3). Surprisingly, spi-b∆232 mutant fish developed normally to adulthood without obvious myeloid phenotypes (data not shown). To further confirm that loss of Spi-b function indeed had no effect on the VDA-derived myeloid development, we outcrossed spi-b∆232 mutants with the Tg(hsp70:mCherry-T2a-CreERT2;mpeg1:LRLG) reporter line and examined the generation of VDA-derived macrophages using IR-LEGO-CreER-loxP labeling. Results showed that spi-b∆232 mutants produced comparable numbers of VDA-derived macrophages to siblings (Figure 2D-F). Likewise, spi-b∆232 mutants contained comparable VDA-derived neutrophils (Figure 2G-H). These results demonstrate that inactivating Spi-b has no effect on the VDA-born myeloid cell development.
Disruption of Spi-1l, the zebrafish ortholog of mammalian SPI-B, does not affect the development of VDA-derived macrophages. (A) Phylogenic tree analysis of mouse SPI genes (PU.1, SPI-B, and SPI-C) with zebrafish counterparts. The phylogenic tree was generated by multiple protein sequence alignment with Clustal Omega online software. (B) Percent identity matrix was created by multiple protein sequence of mammalian SPI-B and zebrafish Pu.1, Spi-b, and Spi-c. (C) Alignment of N-terminal transactivation domain of mouse PU.1 (mPU.1), SPI-B (mSPI-B), and zebrafish Pu.1 (zPu.1) and Spi-1-like (zSpi-1l). Red letters represent the conserved amino acids in acidic motifs (D/E-X-D/E-X-X-X-D/E, blue dashed boxes) and the Q-rich region (purple dashed box), whereas underlined black letters indicate the mutated residues in these position. The black dashed box indicates the P/S/T region in mSPI-B and zSpi-1l, and the P/S/T residues (31.8% in mSPI-B and 37.5% in zSpi-1l) are highlighted by the yellow background. (D-E) Fluorescence imaging of VDA-labeled mpeg1-GFP+ and unlabeled mpeg1-DsRed+ macrophages on the trunk of 8-dpf siblings (D) and spi-b∆232 mutants (E). (F) Quantification of VDA-labeled macrophages (GFP+) in the total macrophage population (mpeg1+). n.s., not significant, Student t test; GFP+/mpeg1+sib (mean/standard error [SE]/n) = 0.40/0.02/6, GFP+/mpeg1+spi-b∆232 (mean/SE/n) = 0.44/0.03/4). Error bars represent SE. (G-H) Sudan Black B (SB) staining of 4 dpf siblings and spi-b∆232 mutants.
Disruption of Spi-1l, the zebrafish ortholog of mammalian SPI-B, does not affect the development of VDA-derived macrophages. (A) Phylogenic tree analysis of mouse SPI genes (PU.1, SPI-B, and SPI-C) with zebrafish counterparts. The phylogenic tree was generated by multiple protein sequence alignment with Clustal Omega online software. (B) Percent identity matrix was created by multiple protein sequence of mammalian SPI-B and zebrafish Pu.1, Spi-b, and Spi-c. (C) Alignment of N-terminal transactivation domain of mouse PU.1 (mPU.1), SPI-B (mSPI-B), and zebrafish Pu.1 (zPu.1) and Spi-1-like (zSpi-1l). Red letters represent the conserved amino acids in acidic motifs (D/E-X-D/E-X-X-X-D/E, blue dashed boxes) and the Q-rich region (purple dashed box), whereas underlined black letters indicate the mutated residues in these position. The black dashed box indicates the P/S/T region in mSPI-B and zSpi-1l, and the P/S/T residues (31.8% in mSPI-B and 37.5% in zSpi-1l) are highlighted by the yellow background. (D-E) Fluorescence imaging of VDA-labeled mpeg1-GFP+ and unlabeled mpeg1-DsRed+ macrophages on the trunk of 8-dpf siblings (D) and spi-b∆232 mutants (E). (F) Quantification of VDA-labeled macrophages (GFP+) in the total macrophage population (mpeg1+). n.s., not significant, Student t test; GFP+/mpeg1+sib (mean/standard error [SE]/n) = 0.40/0.02/6, GFP+/mpeg1+spi-b∆232 (mean/SE/n) = 0.44/0.03/4). Error bars represent SE. (G-H) Sudan Black B (SB) staining of 4 dpf siblings and spi-b∆232 mutants.
The above observations suggest that Pu.1 and Spi-b may play complementary roles in VDA-born myeloid cell development. We therefore examined VDA-born macrophages and neutrophils in pu.1Δ839spi-b∆232 double mutants. We found that, in contrast to pu.1Δ839 and spi-b∆232 single mutants, macrophages derived from both origins were severely impaired in pu.1Δ839spi-b∆232 double mutants as indicated by the lack of mpeg1+ cells from 3 dpf onwards (Figure 3B-H), and by 15 dpf, only a few mpeg1+ macrophages were observed in pu.1Δ839spi-b∆232 double mutants (Figure 3F-H). The depletion of macrophages in pu.1Δ839spi-b∆232 double mutants was attributed to excessive apoptosis of macrophages as indicated by the increase of TUNEL+/coro1a-GFP+ cells in the VDA and caudal hematopoietic tissue (CHT) of 4-dpf double mutants (Figure 3I-J) and AO+mpeg1+ macrophages in the trunk of 15-dpf double mutants (Figure 3K-L). The residual mpeg1+ cells found in pu.1Δ839spi-b∆232 double mutants displayed a small-rounded cell shape with very few branches (Figure 3F′-3G′), suggesting a severe impairment of macrophage differentiation in pu.1Δ839spi-b∆232 double mutants. Likewise, microglia, the central nervous system–resident macrophages were completely absent in pu.1Δ839spi-b∆232 double mutants (Figure 3M-N). However, VDA-born hematopoietic stem/progenitor cells (cmyb+ cells), neutrophils (SB+ cells), and T lymphocytes (rag1+ cells) developed normally (supplemental Figure 4). These results demonstrate that Pu.1 and Spi-b are essential for both the RBI- and VDA-born macrophage development. Notably, the double mutants could not survive to adulthood and died at ∼25 to 30 dpf, perhaps due to the lack of macrophages (data not shown).
Inactivation of Pu.1 and Spi-b impairs the VDA-born macrophage development. (A) A schematic view of a zebrafish embryo. The red box represents the trunk region, the purple box represents CHT, and the blue line indicates the imaged cross section of brain. (B-G) Fluorescence imaging indicates macrophages in Tg(mpeg1:LRLG);pu.1∆839 single and Tg(mpeg1:LRLG);pu.1∆839;spi-bΔ232 double mutants at indicated developmental stages. (F′-G′) Magnified images of the boxed regions as indicated in F and G. (H) Quantification of tissue macrophages in the trunk and CHT (as shown in panels B-G) of Tg(mpeg1:LRLG);pu.1∆839 single and Tg(mpeg1:LRLG);pu.1∆839;spi-bΔ232 double mutants at indicated developmental stages. n.s., not significant; ***P < .001, Student t test; mpeg1+pu.1∆839 (3 dpf, mean/standard error [SE]/n) = 0.6/0.4/7, mpeg1+dmut (3 dpf, mean/SE/n) = 0/0/4, mpeg1+pu.1∆839 (8 dpf, mean/SE/n) = 29.7/1.6/6, mpeg1+dmut (8 dpf, mean/SE/n) = 5.0/1.6/6, mpeg1+pu.1Δ839 (15 dpf, mean/SE/n) = 64.0/5.6/6, mpeg1+dmut (8 dpf, mean/SE/n) = 24.8/2.9/6). Error bars represent SE. (I-J) Costaining of TUNEL signals and coro1a-GFP in the CHT of 4 dpf pu.1∆839 single and pu.1∆839;spi-bΔ232 double mutants. (K-L) Co-staining of acridine orange (AO) signals (K′-L′, green) and Tgmpeg1 (K′′- L′′, red) on the trunk of 15-dpf Tg(mpeg1:LRLG);pu.1Δ839 single and Tg(mpeg1:LRLG);pu.1Δ839;spi-b∆232 double mutants. White arrows in panel L indicate mpeg1+AO+ apoptotic cell bodies of dying macrophages in Tg(mpeg1:LRLG);pu.1Δ839;spi-b∆232 double mutants. (M-N) Costaining of Lcp1 antibody and mpeg1-DsRed on the whole-mount brain sections of pu.1Δ839 mutants and pu.1Δ839;spi-b∆232 double mutants at 15 dpf. Dashed circles indicate the brain regions on the sections.
Inactivation of Pu.1 and Spi-b impairs the VDA-born macrophage development. (A) A schematic view of a zebrafish embryo. The red box represents the trunk region, the purple box represents CHT, and the blue line indicates the imaged cross section of brain. (B-G) Fluorescence imaging indicates macrophages in Tg(mpeg1:LRLG);pu.1∆839 single and Tg(mpeg1:LRLG);pu.1∆839;spi-bΔ232 double mutants at indicated developmental stages. (F′-G′) Magnified images of the boxed regions as indicated in F and G. (H) Quantification of tissue macrophages in the trunk and CHT (as shown in panels B-G) of Tg(mpeg1:LRLG);pu.1∆839 single and Tg(mpeg1:LRLG);pu.1∆839;spi-bΔ232 double mutants at indicated developmental stages. n.s., not significant; ***P < .001, Student t test; mpeg1+pu.1∆839 (3 dpf, mean/standard error [SE]/n) = 0.6/0.4/7, mpeg1+dmut (3 dpf, mean/SE/n) = 0/0/4, mpeg1+pu.1∆839 (8 dpf, mean/SE/n) = 29.7/1.6/6, mpeg1+dmut (8 dpf, mean/SE/n) = 5.0/1.6/6, mpeg1+pu.1Δ839 (15 dpf, mean/SE/n) = 64.0/5.6/6, mpeg1+dmut (8 dpf, mean/SE/n) = 24.8/2.9/6). Error bars represent SE. (I-J) Costaining of TUNEL signals and coro1a-GFP in the CHT of 4 dpf pu.1∆839 single and pu.1∆839;spi-bΔ232 double mutants. (K-L) Co-staining of acridine orange (AO) signals (K′-L′, green) and Tgmpeg1 (K′′- L′′, red) on the trunk of 15-dpf Tg(mpeg1:LRLG);pu.1Δ839 single and Tg(mpeg1:LRLG);pu.1Δ839;spi-b∆232 double mutants. White arrows in panel L indicate mpeg1+AO+ apoptotic cell bodies of dying macrophages in Tg(mpeg1:LRLG);pu.1Δ839;spi-b∆232 double mutants. (M-N) Costaining of Lcp1 antibody and mpeg1-DsRed on the whole-mount brain sections of pu.1Δ839 mutants and pu.1Δ839;spi-b∆232 double mutants at 15 dpf. Dashed circles indicate the brain regions on the sections.
Pu.1 and Spi-b form a parallel genetic network to regulate VDA-born macrophage development
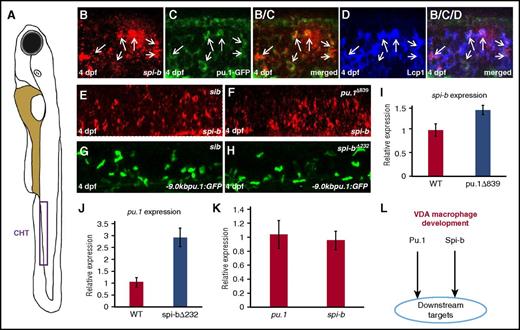
To dissect the genetic hierarchy of pu.1 and spi-b in the VDA-born macrophage development, we first performed spi-b RNA WISH and anti-GFP antibody double staining to examine the expression of pu.1 and spi-b in Tg(pu.1:GFP) transgenic fish28 at 4 dpf, when VDA-born hematopoietic cells are known to colonize the CHT.40,41 Results showed that majority of the pu.1-GFP cells in the CHT were positive for spi-b mRNA (Figure 4B-D), showing that pu.1 and spi-b are co-expressed in the same VDA-born myeloid cells. Thus, Pu.1 and Spi-b appear to act in parallel to regulate the development of VDA-derived macrophages. To support this notion, we examined the expression of spi-b and pu.1 by monitoring spi-b transcripts and GFP signal in the CHT region of pu.1Δ839 mutants and spi-b∆232;Tg(-9.0kbpu.1:eGFP) mutants. As indicated in Figure 4E-H, spi-b and pu.1 transcripts were clearly not downregulated in pu.1Δ839 and spi-b∆232;Tg(-9.0kbpu.1:eGFP) mutants, respectively (Figure 4E-H). In fact, real-time RT-PCR analysis with VDA-derived macrophages isolated from pu.1 MO-injected wild-type Tg(mpeg1:LRLG) (pu.1 MO blocks the development of RBI-, but not VDA-derived, macrophages; supplemental Figure 5), pu.1Δ839;Tg(mpeg1:LRLG), and spi-b∆232;Tg(mpeg1:LRLG) mutants revealed an upregulation of spi-b and pu.1 transcripts in pu.1Δ839 and spi-b∆232 mutants, respectively, perhaps due to the compensation effect (Figure 4I-J). Consistent with the genetic results that pu.1 and spi-b are capable of fully compensating each other, pu.1 and spi-b were expressed at comparable levels in the VDA-derived macrophages (Figure 4K). Taken together, we conclude that pu.1 and spi-b act in parallel and cooperatively to regulate the VDA-derived macrophage development (Figure 4L).
Pu.1 and Spi-b function in parallel during VDA-derived macrophage development. (A) Schematic view of a zebrafish embryo. Purple box indicates the CHT. (B-D) Costaining of spi-b mRNA with Tg(-9.0kbpu.1:GFP) and Lcp1 antibody in the CHT region of 4-dpf wild-type embryos. Panels B/C and B/C/D are merged images from the corresponding panels (B-D). Arrows in each panel indicate cells coexpressing spi-b, pu.1-GFP, and Lcp1. (E-F) WISH of spi-b expression in 4-dpf siblings and pu.1∆839 mutants. (G-H) Fluorescence images of the pu.1-GFP+ myeloid cells in the CHT of 4-dpf Tg(-9.0kbpu.1:GFP) siblings and Tg(-9.0kbpu.1:GFP);spi-b∆232 mutants. (I) Quantitative RT-PCR for spi-b expression in VDA-derived macrophages isolated from pu.1 MO injected wild-type Tg(mpeg1:LRLG) and pu.1∆839;Tg(mpeg1:LRLG) at 8 dpf. (J) Quantitative RT-PCR for pu.1 expression in the VDA-derived macrophages isolated from pu.1 MO-injected wild-type Tg(mpeg1:LRLG) and spi-b∆232;Tg(mpeg1:LRLG) at 8 dpf. (K) Quantitative RT-PCR for pu.1 and spi-b expression in VDA-derived macrophages isolated from pu.1 MO-injected Tg(mpeg1:LRLG) at 8 dpf. Expression level of target genes in panels I-K was normalized with elf1a expression. Error bars represent standard error. (L) The parallel Pu.1−Spi-b genetic network in the development of VDA-derived macrophages.
Pu.1 and Spi-b function in parallel during VDA-derived macrophage development. (A) Schematic view of a zebrafish embryo. Purple box indicates the CHT. (B-D) Costaining of spi-b mRNA with Tg(-9.0kbpu.1:GFP) and Lcp1 antibody in the CHT region of 4-dpf wild-type embryos. Panels B/C and B/C/D are merged images from the corresponding panels (B-D). Arrows in each panel indicate cells coexpressing spi-b, pu.1-GFP, and Lcp1. (E-F) WISH of spi-b expression in 4-dpf siblings and pu.1∆839 mutants. (G-H) Fluorescence images of the pu.1-GFP+ myeloid cells in the CHT of 4-dpf Tg(-9.0kbpu.1:GFP) siblings and Tg(-9.0kbpu.1:GFP);spi-b∆232 mutants. (I) Quantitative RT-PCR for spi-b expression in VDA-derived macrophages isolated from pu.1 MO injected wild-type Tg(mpeg1:LRLG) and pu.1∆839;Tg(mpeg1:LRLG) at 8 dpf. (J) Quantitative RT-PCR for pu.1 expression in the VDA-derived macrophages isolated from pu.1 MO-injected wild-type Tg(mpeg1:LRLG) and spi-b∆232;Tg(mpeg1:LRLG) at 8 dpf. (K) Quantitative RT-PCR for pu.1 and spi-b expression in VDA-derived macrophages isolated from pu.1 MO-injected Tg(mpeg1:LRLG) at 8 dpf. Expression level of target genes in panels I-K was normalized with elf1a expression. Error bars represent standard error. (L) The parallel Pu.1−Spi-b genetic network in the development of VDA-derived macrophages.
Spi-b is involved in and partially compensates for Pu.1 function during RBI-born macrophage development
A previous study reported by Bukrinsky et al showed that the suppression of Spi-b function by spi-b MO severely blocks embryonic (RBI-born) myeloid cell development.26 We were therefore keen to find out whether RBI-derived myeloid cells are impaired in spi-b∆232 mutants. Surprisingly, we found that RBI-born embryonic neutrophils and myeloid progenitors, peripheral macrophages, and microglia in the brain developed normally in spi-b∆232 mutants (Figure 5A-H). This result is somewhat unexpected, as spi-b is highly expressed in RBI-derived myeloid cells during early myelopoiesis26 (Figure 6C). We hypothesized that because of the compensation of Pu.1, the role of Spi-b in the RBI-born myeloid cell development could not be revealed in spi-b–deficient mutants. To test this hypothesis, we used a weak hypomorphic pu.1 allele, pu.1∆371, which harbors an in-frame 42-amino-acid deletion in the transactivation domain, resulting in the synthesis of a truncated Pu.1 protein with reduced transactivation activity (supplemental Figure 6A-D). In these weak hypomorphic mutants, RBI-derived macrophages and microglia were only partially reduced (supplemental Figure 6E-H). We then outcrossed pu.1Δ371 mutants with spi-b∆232 and asked if inactivating Spi-b in this pu.1Δ371 hypomorphic mutant background would cause a more severe phenotype. Indeed, the number of RBI-derived macrophages (Figure 5I-J,M), but not neutrophils (Figure 5K-M), was further reduced in pu.1Δ371;spi-b∆232 double mutants. These results demonstrate that Spi-b participates in RBI-derived macrophage development, although its role is limited.
Spi-b partially compensates for Pu.1 function during the development of RBI-born macrophages. (A-B) WISH of mfap4 expression in 30-hpf siblings (35/35) and spi-b∆232 mutants (n = 14/14). (C-D) Neutral red staining of microglia in 3-dpf siblings and spi-b∆232 mutants. (E-F) WISH of lyz expression in 30-hpf siblings (n = 38/38) and spi-b∆232 mutants (n = 15/15). (G-H) WISH of pu.1 expression in 20-hpf siblings (n = 44/44) and spi-b∆232 mutants (n = 17/17). (I-J) WISH of mfap4 expression in 36-hpf pu.1Δ371 single- and pu.1Δ371;spi-b∆232 double-mutant embryos. (K-L) WISH of lyz expression in 36-hpf pu.1Δ371 single- and pu.1Δ371;spi-b∆232 double-mutant embryos. (M) Quantification of mfap4+ macrophages and lyz+ neutrophils as shown in panels I-J and K-L, respectively. n.s., not significant, ***P < .001, Student t test; mfap4+pu.1Δ371 (mean/standard error [SE]/n) = 39.0/3.0/16, mfap4+dmut (mean/SE/n) = 22.9/2.2/18, lyz+pu.1Δ371 (mean/SE/n)=117.4/4.4/7, lyz+dmut (mean/SE/n) =121.6/4.2/8). Errors bars represent SE.
Spi-b partially compensates for Pu.1 function during the development of RBI-born macrophages. (A-B) WISH of mfap4 expression in 30-hpf siblings (35/35) and spi-b∆232 mutants (n = 14/14). (C-D) Neutral red staining of microglia in 3-dpf siblings and spi-b∆232 mutants. (E-F) WISH of lyz expression in 30-hpf siblings (n = 38/38) and spi-b∆232 mutants (n = 15/15). (G-H) WISH of pu.1 expression in 20-hpf siblings (n = 44/44) and spi-b∆232 mutants (n = 17/17). (I-J) WISH of mfap4 expression in 36-hpf pu.1Δ371 single- and pu.1Δ371;spi-b∆232 double-mutant embryos. (K-L) WISH of lyz expression in 36-hpf pu.1Δ371 single- and pu.1Δ371;spi-b∆232 double-mutant embryos. (M) Quantification of mfap4+ macrophages and lyz+ neutrophils as shown in panels I-J and K-L, respectively. n.s., not significant, ***P < .001, Student t test; mfap4+pu.1Δ371 (mean/standard error [SE]/n) = 39.0/3.0/16, mfap4+dmut (mean/SE/n) = 22.9/2.2/18, lyz+pu.1Δ371 (mean/SE/n)=117.4/4.4/7, lyz+dmut (mean/SE/n) =121.6/4.2/8). Errors bars represent SE.
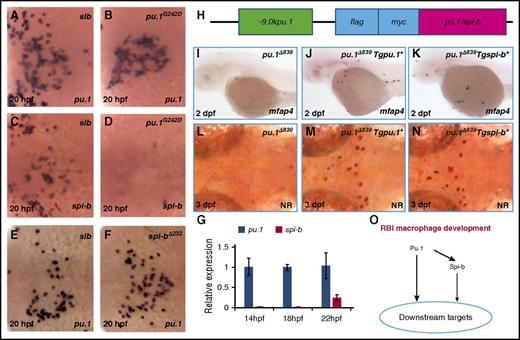
The formation of RBI-born macrophages is regulated by a vertical Pu.1−Spi-b axis. (A-B) WISH of pu.1 expression in 20-hpf siblings (n = 39/39) and hypomorphic pu.1G242D mutants (n = 13/13). (C-D) WISH of spi-b expression in 20-hpf siblings (n = 41/41) and pu.1G242D mutants (n = 12/12). (E-F) WISH of pu.1 expression in 20-hpf siblings (n = 45/45) and spi-b∆232 mutants (n = 16/16). All embryos in panels A-F are dorsally orientated with anterior to the left. (G) Quantitative RT-PCR for pu.1 and spi-b expression in wild-type embryos at indicated stages (x-axis). Expression level of target genes was normalized with elf1a expression. Error bars represent standard error. (H) Schematic view of the pu.1 or spi-b expression constructs under the control of a -9.0kb pu.1 promoter. Both Pu.1 and Spi-b are flag-myc double tagged at the N terminus. (I-K) WISH of mfap4 expression in 2-dpf pu.1∆839 (n = 25/25), pu.1∆839;Tg(-9.0kbpu.1:flag-myc-pu.1)+ (shorted as Tgpu.1+) (n = 14/15), and pu.1∆839;Tg(-9.0kbpu.1:flag-myc-spi-b)+ (shorted as Tgspi-b+) (n = 11/13). (L-N) Neutral red staining of microglia in 3-dpf pu.1∆839, pu.1∆839;Tgpu.1+, and pu.1∆839;Tgspi-b+. (O) The working model for the vertical Pu.1−Spi-b genetic network in RBI macrophage development.
The formation of RBI-born macrophages is regulated by a vertical Pu.1−Spi-b axis. (A-B) WISH of pu.1 expression in 20-hpf siblings (n = 39/39) and hypomorphic pu.1G242D mutants (n = 13/13). (C-D) WISH of spi-b expression in 20-hpf siblings (n = 41/41) and pu.1G242D mutants (n = 12/12). (E-F) WISH of pu.1 expression in 20-hpf siblings (n = 45/45) and spi-b∆232 mutants (n = 16/16). All embryos in panels A-F are dorsally orientated with anterior to the left. (G) Quantitative RT-PCR for pu.1 and spi-b expression in wild-type embryos at indicated stages (x-axis). Expression level of target genes was normalized with elf1a expression. Error bars represent standard error. (H) Schematic view of the pu.1 or spi-b expression constructs under the control of a -9.0kb pu.1 promoter. Both Pu.1 and Spi-b are flag-myc double tagged at the N terminus. (I-K) WISH of mfap4 expression in 2-dpf pu.1∆839 (n = 25/25), pu.1∆839;Tg(-9.0kbpu.1:flag-myc-pu.1)+ (shorted as Tgpu.1+) (n = 14/15), and pu.1∆839;Tg(-9.0kbpu.1:flag-myc-spi-b)+ (shorted as Tgspi-b+) (n = 11/13). (L-N) Neutral red staining of microglia in 3-dpf pu.1∆839, pu.1∆839;Tgpu.1+, and pu.1∆839;Tgspi-b+. (O) The working model for the vertical Pu.1−Spi-b genetic network in RBI macrophage development.
Pu.1 and Spi-b form a vertical genetic axis to control RBI-born macrophage development
To delineate the molecular basis underlying the differential requirements of Pu.1 and Spi-b for RBI-derived macrophage development, we examined the expression of spi-b and pu.1 in RBI-derived myeloid cells in hypomorphic pu.1G242D mutants and spi-b∆232 mutants. WISH showed that the expression of spi-b was completely abolished in pu.1G242D mutants (Figure 6C-D), despite the presence of RBI-derived myeloid progenitors (Figure 6A-B). In contrast, the pu.1 transcripts were intact in spi-b∆232 mutants (Figure 6E-F). These results indicate that spi-b is genetically downstream of Pu.1 during embryonic myeloid cell development. Interestingly, real-time RT-PCR analysis revealed that the expression level of spi-b in embryonic myeloid cells was significantly lower than that of pu.1 (Figure 6G), indicating that the limited role of Spi-b in RBI-derived macrophage development is due to its low expression. Indeed, when expressed at a similar level using a previously characterized pu.1 promoter,28 the ability of Spi-b or Pu.1 to rescue RBI-derived macrophages was indistinguishable in pu.1Δ839 mutants (Figure 6H-N). Taken together, we conclude that during RBI-derived macrophage development, Pu.1 is a master regulator that acts upstream of Spi-b, which, upon induction by Pu.1, can partially compensate for the function of Pu.1 (Figure 6O).
The distinct Pu.1-Spi-b regulatory networks govern macrophage development partially through regulating Irf8 expression
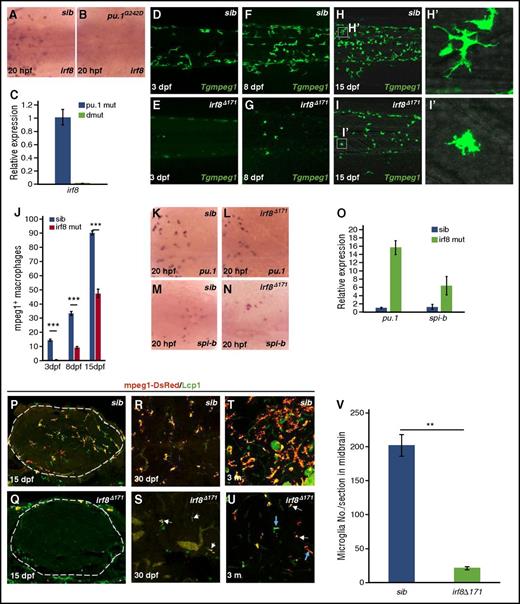
The above genetic analysis has shown that pu.1 and spi-b form 2 distinct genetic networks, a vertical and a parallel cascade, to regulate RBI- and VDA-born macrophages, respectively. To identify the key targets downstream of these distinct Pu.1−Spi-b regulatory networks, we focused on Irf8, a transcription factor known to be a Pu.1 downstream target and essential for macrophage development.34,42,43 Indeed, WISH showed that the irf8 expression in RBI-born myeloid cells was completely absent in pu.1-deficient mutants (both hypomorphic pu.1G242D and null pu.1Δ839 alleles) (Figure 7A-B). Likewise, real-time RT-PCR analysis with the residual VDA-derived macrophages isolated from 20-dpf Tg(mpeg1:LRLG);pu.1Δ839;spi-bΔ232 double mutants showed that the irf8 expression was also abolished in the VDA-born macrophages in pu.1Δ839;spi-bΔ232 mutants (Figure 7C). To confirm that irf8 is a key downstream target of both the vertical and the parallel Pu.1−Spi-b regulatory cascades, we generated an irf8-null allele, irf8Δ171, which harbors a 171-bp deletion and a 3-bp insertion in exon 1, resulting in the generation of a truncated protein lacking the DNA-binding domain due to the usage of the downstream ATG for translation initiation (supplemental Figure 7). Similar to pu.1Δ839;spi-bΔ232 double mutants, RBI- and VDA-born macrophages (peripheral macrophages and microglia) in irf8Δ171 mutants were drastically reduced or absent (Figure 7D-I,P-Q). As expected, the expression of pu.1 and spi-b in RBI- and VDA-derived myeloid cells remained either unchanged or upregulated in irf8Δ171 mutants (Figure 7K-O), perhaps due to a feedback mechanism. These data demonstrate that the vertical and parallel Pu.1−Spi-b regulatory networks orchestrate the development of RBI- and VDA-derived macrophages, at least in part, by regulating a common downstream regulator, Irf8. Notably, in contrast to the complete absence of microglia and early lethality of pu.1Δ839;spi-bΔ232 double mutants, irf8Δ171 mutant fish are viable and have a partial recovery of mpeg1+ cells in the brain in adulthood (Figure 7R-V). The recovered mpeg1+ cells in adult irf8Δ171 mutants appeared to be macrophages, as they lacked Mpx activity, a typical feature of neutrophils (supplemental Figure 8C-D), and were able to engulf bacteria (supplemental Figure 8A-B). This observation suggests that the development of a small portion of VDA-born macrophages is independent of Irf8.
The paralleled and vertical Pu.1−Spi-b cascades control macrophage development via a common downstream factor, Irf8. (A-B) WISH of irf8 expression in 20-hpf siblings (n = 35/35) and hypomorphic pu.1G242D mutants (n = 11/11). (C) Quantitative RT-PCR for irf8 expression in purified macrophages isolated from pu.1∆839;Tg(mpeg1:LRLG) and pu.1Δ839;spi-b∆232;Tg(mpeg1:LRLG) at 20 dpf. Expression level of target genes was normalized with elf1a expression. Error bars represent standard error (SE). (D-I) Fluorescence imaging of peripheral macrophages in the trunk and CHT regions of Tg(mpeg1:LRLG) siblings and Tg(mpeg1:LRLG);irf8∆171 mutants at indicated developmental stages. (H′-I′) Magnified images of the boxed regions as indicated in panels H and I. (J) Quantification of tissue macrophages in the trunk and CHT regions (as shown in panels D-I) Tg(mpeg1:LRLG) siblings and Tg(mpeg1:LRLG);irf8∆171 mutants at indicated developmental stages. ***P < .001, Student t test; mpeg1+sib (3 dpf, mean/SE/n)= 14.3/15/0.7, mpeg1+irf8 mut (3 dpf, mean/SE/n) = 0.5/15/0.2, mpeg1+sib (8 dpf, mean/SE/n) = 33.3/10/1.3, mpeg1+irf8 mut (8 dpf, mean/SE/n) = 9.1/10/0.7, mpeg1+sib(15 dpf, mean/SE/n) = 90.2/10/1.4, mpeg1+irf8 mut (15 dpf, mean/SE/n) = 47.3/12/3.1). Error bars represent SE. (K-N) WISH of pu.1 and spi-b expression in 20-hpf siblings (pu.1, n = 40/40; spi-b, n = 38/38) and irf8∆171 mutants (pu.1, n = 15/15; spi-b, n = 12/12). (O) Quantitative RT-PCR for pu.1 and spi-b expression in purified macrophages isolated from wild-type Tg(mpeg1:LRLG) and irf8∆171;Tg(mpeg1:LRLG) at 20 dpf. Expression level of target genes was normalized with elf1a expression. Error bars represent SE. (P-Q) Costaining of Lcp1 antibody and mpeg1-DsRed on whole-mount brain sections of 15-dpf Tg(mpeg1:LRLG) siblings and Tg(mpeg1:LRLG);irf8∆171 mutants. Dashed circles indicate the brain region. (R-S) Costaining of Lcp1 antibody and mpeg1-DsRed on brain sections of Tg(mpeg1:LRLG) siblings and Tg(mpeg1:LRLG);irf8∆171 mutants at 30 dpf. White arrows in panel S indicate mpeg1-DsRed+/Lcp1+ microglia in irf8∆171 mutants. (T-U) Costaining of Lcp1 antibody and mpeg1-DsRed on the brain sections of adult (3-month-old) Tg(mpeg1:LRLG) siblings and Tg(mpeg1:LRLG);irf8∆171 mutants. Blue and white arrows in panel U indicate branched microglia and rounded microglia-like cells in irf8∆171 mutants, respectively. Images are selected as representatives. (V) Quantification of Lcp1 and mpeg1-DsRed double-positive microglia in the midbrain of 3-month-old wild-type siblings and irf8∆171 mutants. **P < .01, Student t test; sib (mean/SE/n) = 201.7/15.8/4, irf8∆171 (mean/SE/n) = 21.4/2.2/3). Error bars represent SE.
The paralleled and vertical Pu.1−Spi-b cascades control macrophage development via a common downstream factor, Irf8. (A-B) WISH of irf8 expression in 20-hpf siblings (n = 35/35) and hypomorphic pu.1G242D mutants (n = 11/11). (C) Quantitative RT-PCR for irf8 expression in purified macrophages isolated from pu.1∆839;Tg(mpeg1:LRLG) and pu.1Δ839;spi-b∆232;Tg(mpeg1:LRLG) at 20 dpf. Expression level of target genes was normalized with elf1a expression. Error bars represent standard error (SE). (D-I) Fluorescence imaging of peripheral macrophages in the trunk and CHT regions of Tg(mpeg1:LRLG) siblings and Tg(mpeg1:LRLG);irf8∆171 mutants at indicated developmental stages. (H′-I′) Magnified images of the boxed regions as indicated in panels H and I. (J) Quantification of tissue macrophages in the trunk and CHT regions (as shown in panels D-I) Tg(mpeg1:LRLG) siblings and Tg(mpeg1:LRLG);irf8∆171 mutants at indicated developmental stages. ***P < .001, Student t test; mpeg1+sib (3 dpf, mean/SE/n)= 14.3/15/0.7, mpeg1+irf8 mut (3 dpf, mean/SE/n) = 0.5/15/0.2, mpeg1+sib (8 dpf, mean/SE/n) = 33.3/10/1.3, mpeg1+irf8 mut (8 dpf, mean/SE/n) = 9.1/10/0.7, mpeg1+sib(15 dpf, mean/SE/n) = 90.2/10/1.4, mpeg1+irf8 mut (15 dpf, mean/SE/n) = 47.3/12/3.1). Error bars represent SE. (K-N) WISH of pu.1 and spi-b expression in 20-hpf siblings (pu.1, n = 40/40; spi-b, n = 38/38) and irf8∆171 mutants (pu.1, n = 15/15; spi-b, n = 12/12). (O) Quantitative RT-PCR for pu.1 and spi-b expression in purified macrophages isolated from wild-type Tg(mpeg1:LRLG) and irf8∆171;Tg(mpeg1:LRLG) at 20 dpf. Expression level of target genes was normalized with elf1a expression. Error bars represent SE. (P-Q) Costaining of Lcp1 antibody and mpeg1-DsRed on whole-mount brain sections of 15-dpf Tg(mpeg1:LRLG) siblings and Tg(mpeg1:LRLG);irf8∆171 mutants. Dashed circles indicate the brain region. (R-S) Costaining of Lcp1 antibody and mpeg1-DsRed on brain sections of Tg(mpeg1:LRLG) siblings and Tg(mpeg1:LRLG);irf8∆171 mutants at 30 dpf. White arrows in panel S indicate mpeg1-DsRed+/Lcp1+ microglia in irf8∆171 mutants. (T-U) Costaining of Lcp1 antibody and mpeg1-DsRed on the brain sections of adult (3-month-old) Tg(mpeg1:LRLG) siblings and Tg(mpeg1:LRLG);irf8∆171 mutants. Blue and white arrows in panel U indicate branched microglia and rounded microglia-like cells in irf8∆171 mutants, respectively. Images are selected as representatives. (V) Quantification of Lcp1 and mpeg1-DsRed double-positive microglia in the midbrain of 3-month-old wild-type siblings and irf8∆171 mutants. **P < .01, Student t test; sib (mean/SE/n) = 201.7/15.8/4, irf8∆171 (mean/SE/n) = 21.4/2.2/3). Error bars represent SE.
Discussion
In this report, by combining genetics and a temporal-spatial–resolution fate-mapping method, we reveal that the development of macrophages of different origins is orchestrated by distinct genetic networks comprising the transcription factors Pu.1, Spi-b, and Irf8.
Our study has shown that genetic inactivation of Spi-b does not affect the development of both RBI- and VDA-born myeloid cells. This result is clearly different from those of a previous study, where Bukrinsky et al showed that transient knockdown of spi-b expression by the spi-b MO severely impairs the development of embryonic or RBI-born myeloid cells.26 Three possible mechanisms could explain this discrepancy. One explanation is that the discrepancy is caused by the toxic and off-target effects of MO44 or, alternatively, the difference is attributed to the complementary effect that occurs only in the genetic spi-b mutants.45 Finally, although the majority of the Spi-b Ets domain was removed in spi-bΔ232 mutants, we cannot exclude the possibility that weak Spi-b activity was retained, which might be sufficient to sustain the development of RBI-derived macrophages.
Our findings challenge the current view that zebrafish Pu.1 and Spi1-like are co-orthologs of mammalian PU.1. Two lines of evidence support the notion that Spi1-like is the ortholog of mammalian SPI-B.46 The first is provided by protein sequence alignment and phylogenic tree analysis, which reveals that Spi1-like shares the highest similarity with mammalian SPI-B among Ets family transcription factors and the transactivation domain of Spi1-like mimics that of mammalian SPI-B, but not PU.1. The second line of evidence is provided by functional studies showing that similar to spi-bΔ232 mutant zebrafish, SPI-B knockout mice are viable, with no obvious hematopoietic lineage developmental defects,23 and SPI-B acts cooperatively with PU.1 and can compensate for the function of PU.1 in hematopoietic cell development.47-49
Our study demonstrates that RBI- and VDA-derived macrophages are regulated by distinct genetic networks formed by Pu.1 and Spi-b. Although the establishment of these distinct genetic networks remains unknown, the parallel Pu.1−Spi-b cascade does give a competitive advantage as a single copy of either gene is sufficient to support the development of macrophages and ensure the survival of the organism. Unexpectedly, both vertical and parallel Pu.1−Spi-b cascades govern the macrophage development through, at least in part, regulating a common key downstream target irf8. Notably, a recent study in mice has reported that during dendritic cell development PU.1 directly binds the distal enhancers (−50 kb) of the IRF8 locus, leading to the activation of IRF8 transcription.50 Remarkably, promoter analysis reveals 2 discrete clusters of potential Pu.1 binding sites at the −16-kb to −14-kb and −10.8-kb to −8.8-kb regions of the zebrafish irf8 locus (supplemental Figure 9), suggesting a conserved regulatory mechanism across species. However, forced expression of irf8 using the hsp70 heat shock promoter is insufficient to restore the development of either RBI-derived macrophages in pu.1Δ839 single34 or VDA-derived macrophages in pu.1Δ839;spi-bΔ232 double mutants (data not shown), suggesting that other transcription factors that act in parallel with Irf8 are involved in macrophage development. This notion is supported by the finding that macrophage defects in irf8-null mutants are less severe than those seen in pu.1Δ839;spi-bΔ232 double mutants and by the finding that a small proportion of macrophages continue to develop into microglia in an Irf8-independent fashion. Further studies will be required to uncover these factors.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Michael Brand for sharing the Tg(hsp70:mCherry-T2a-CreERT2) transgenic line, Thomas Look for providing the Tg(zpu.1:eGFP)df5 transgenic line, and Robert Qi for providing anti-GFP antibody.
This work was supported by grants from the National Natural Science Foundation of China (31229003), the Research Grants Council of the Hong Kong Special Administrative region (16102414, 16103515, HKUST5/CRF/12R, AoE/M-09/12, and T13-607/12R), and the Innovation and Technology Commission of the Hong Kong Special Administrative region (ITCPD/17-9)
Authorship
Contribution: T.Y., W.G., Y.T., J.X., and J.C. designed the research, performed experiments, and analyzed data; L.L. generated irf8Δ171 mutants; and Z.W. designed the research and analyzed data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Zilong Wen, Division of Life Science, State Key Laboratory of Molecular Neuroscience and Center of Systems Biology and Human Health, Hong Kong University of Science and Technology, Clear Water Bay, Kowloon, Hong Kong, People’s Republic of China; e-mail: zilong@ust.hk.

![Figure 1. Pu.1 is required for the RBI- but not VDA-derived macrophage development. (A-B) WISH of mfap4 expression in 30-hpf siblings (n = 31/31) and pu.1∆839 mutants (n = 12/12). (C-D) Neutral red staining of microglia in 3-dpf siblings and pu.1∆839 mutants. (E-F) WISH of lyz expression in 30-hpf siblings (n = 35/35) and pu.1∆839 mutants (n = 13/13). (G-H) WISH of cebpa expression in 20-hpf siblings (n = 42/42) and pu.1∆839 mutants (n = 13/13). (I-J) Fluorescence imaging of RBI- and VDA-labeled pu.1 ∆839;Tg(mpeg1:Loxp-Dsred-loxp-GFP) embryos at 8 dpf. Embryos are laterally orientated to view macrophages in the trunk region. (K) Quantification of RBI- or VDA-labeled macrophages (GFP+) in the total recovered macrophages (mpeg1+) in 8 dpf pu.1∆839 mutants compared with control embryos without heat shock. ***P < .001 (Student t test, GFP+/mpeg1+control (mean/standard error [SE]/n) = 0.09/0.01/9), GFP+/mpeg1+RBI (mean/SE/n) = 0.08/0.02/4, GFP+/mpeg1+VDA (mean/SE/n) = 0.66/0.04/7). Error bars represent SE. (L-M) Sudan black B (SB) staining of neutrophils in 4-dpf siblings and pu.1∆839 mutants.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/129/4/10.1182_blood-2016-07-727651/4/m_blood727651f1.jpeg?Expires=1769080560&Signature=1i28BR5glOyB6Dm6LDJAnlOrBm5bbkNgXtqOdN1IQmcm9~MMsc4Y~fYC-X~TPtzmlF36FA6OMD2pT8G92XTkjWw08NmhT72TjBrb4Mstl8R2z2nY-7labTS5YU3ZJ0axedil4mJPWpTxBNaSUqw4fRXls09fW-gw4c5QT0lucZfnc5rzzgWStUfmO0-LDh~0PNR0nw4fQGfS-LgLjjQqa9p0UMa2Rl5t4sqOR2syB-NXKe1y3w-Z2NCqTfmQ45Fjq14ALI7TRtRt3yKd0Oe8oM-nfAiXugvvvyyx4gBywMsSfhAse-0TNmKdZruAyd~T~GVu3EmDyU~jrlUorqf1PA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 2. Disruption of Spi-1l, the zebrafish ortholog of mammalian SPI-B, does not affect the development of VDA-derived macrophages. (A) Phylogenic tree analysis of mouse SPI genes (PU.1, SPI-B, and SPI-C) with zebrafish counterparts. The phylogenic tree was generated by multiple protein sequence alignment with Clustal Omega online software. (B) Percent identity matrix was created by multiple protein sequence of mammalian SPI-B and zebrafish Pu.1, Spi-b, and Spi-c. (C) Alignment of N-terminal transactivation domain of mouse PU.1 (mPU.1), SPI-B (mSPI-B), and zebrafish Pu.1 (zPu.1) and Spi-1-like (zSpi-1l). Red letters represent the conserved amino acids in acidic motifs (D/E-X-D/E-X-X-X-D/E, blue dashed boxes) and the Q-rich region (purple dashed box), whereas underlined black letters indicate the mutated residues in these position. The black dashed box indicates the P/S/T region in mSPI-B and zSpi-1l, and the P/S/T residues (31.8% in mSPI-B and 37.5% in zSpi-1l) are highlighted by the yellow background. (D-E) Fluorescence imaging of VDA-labeled mpeg1-GFP+ and unlabeled mpeg1-DsRed+ macrophages on the trunk of 8-dpf siblings (D) and spi-b∆232 mutants (E). (F) Quantification of VDA-labeled macrophages (GFP+) in the total macrophage population (mpeg1+). n.s., not significant, Student t test; GFP+/mpeg1+sib (mean/standard error [SE]/n) = 0.40/0.02/6, GFP+/mpeg1+spi-b∆232 (mean/SE/n) = 0.44/0.03/4). Error bars represent SE. (G-H) Sudan Black B (SB) staining of 4 dpf siblings and spi-b∆232 mutants.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/129/4/10.1182_blood-2016-07-727651/4/m_blood727651f2.jpeg?Expires=1769080560&Signature=qOvDfApRovCda4AAZ5FU5Or79hGOASqmIL8CC3V6sVGDtc31CZMKg6S4Y6wJTBpgKzQr--DgELvvg1nyUFViuLdDKJmEwOaJKZSNHy0t8S0vE-dFCaH~NJpLh-uhom4pCILeFA1sQxWZi6umjStbO41~6fU6BiWPlLSnvHKoTOWLiieONTuHI5XRXS4lwiFyvywsdbFIXcBCrwOrVWIshn9aWRd6OygS0UwM75GxcYdBeECL7uA0Ox7oI7Fsn~Wm4J15SnlaqAfg494IiEBOo2R6QybbohmpfGZAhgC6EjD0-9IDwi21Xbrigui59tYxyqdGBfU~sbcRUuoA3YOS3g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 3. Inactivation of Pu.1 and Spi-b impairs the VDA-born macrophage development. (A) A schematic view of a zebrafish embryo. The red box represents the trunk region, the purple box represents CHT, and the blue line indicates the imaged cross section of brain. (B-G) Fluorescence imaging indicates macrophages in Tg(mpeg1:LRLG);pu.1∆839 single and Tg(mpeg1:LRLG);pu.1∆839;spi-bΔ232 double mutants at indicated developmental stages. (F′-G′) Magnified images of the boxed regions as indicated in F and G. (H) Quantification of tissue macrophages in the trunk and CHT (as shown in panels B-G) of Tg(mpeg1:LRLG);pu.1∆839 single and Tg(mpeg1:LRLG);pu.1∆839;spi-bΔ232 double mutants at indicated developmental stages. n.s., not significant; ***P < .001, Student t test; mpeg1+pu.1∆839 (3 dpf, mean/standard error [SE]/n) = 0.6/0.4/7, mpeg1+dmut (3 dpf, mean/SE/n) = 0/0/4, mpeg1+pu.1∆839 (8 dpf, mean/SE/n) = 29.7/1.6/6, mpeg1+dmut (8 dpf, mean/SE/n) = 5.0/1.6/6, mpeg1+pu.1Δ839 (15 dpf, mean/SE/n) = 64.0/5.6/6, mpeg1+dmut (8 dpf, mean/SE/n) = 24.8/2.9/6). Error bars represent SE. (I-J) Costaining of TUNEL signals and coro1a-GFP in the CHT of 4 dpf pu.1∆839 single and pu.1∆839;spi-bΔ232 double mutants. (K-L) Co-staining of acridine orange (AO) signals (K′-L′, green) and Tgmpeg1 (K′′- L′′, red) on the trunk of 15-dpf Tg(mpeg1:LRLG);pu.1Δ839 single and Tg(mpeg1:LRLG);pu.1Δ839;spi-b∆232 double mutants. White arrows in panel L indicate mpeg1+AO+ apoptotic cell bodies of dying macrophages in Tg(mpeg1:LRLG);pu.1Δ839;spi-b∆232 double mutants. (M-N) Costaining of Lcp1 antibody and mpeg1-DsRed on the whole-mount brain sections of pu.1Δ839 mutants and pu.1Δ839;spi-b∆232 double mutants at 15 dpf. Dashed circles indicate the brain regions on the sections.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/129/4/10.1182_blood-2016-07-727651/4/m_blood727651f3.jpeg?Expires=1769080560&Signature=Wvd3Kdw88oW-cc74W842h3-0ruTPHLptkcmksIR3tdbvGc-Z7ANEqWAZl5FLbcakU-qcKwdjkM6RDhIyX20GBdWT2F12cvWGkQIoihBVzhUgjsppW~I5l~mNahW2UfRv4IPM9wQwaby3hJAONmwKZsYFBxSEcW9Ov2dtNEe1oJ8HfXmN4~pLpotE8T~2DtV8W37mQg7LHtIztozHp9ueGCm3~Il1D-2Qhrw4qJ8bXG6dQczqWxmfs-0Lix73Z4JV9Ehiq0oab4SYrMjpmkMEjbEBNQB8-NJfyr1KCAiXKrnUxyeAa~0QXY-zCIvbwRgiHzkW5ZQA12JL8UEW9bDWHQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 5. Spi-b partially compensates for Pu.1 function during the development of RBI-born macrophages. (A-B) WISH of mfap4 expression in 30-hpf siblings (35/35) and spi-b∆232 mutants (n = 14/14). (C-D) Neutral red staining of microglia in 3-dpf siblings and spi-b∆232 mutants. (E-F) WISH of lyz expression in 30-hpf siblings (n = 38/38) and spi-b∆232 mutants (n = 15/15). (G-H) WISH of pu.1 expression in 20-hpf siblings (n = 44/44) and spi-b∆232 mutants (n = 17/17). (I-J) WISH of mfap4 expression in 36-hpf pu.1Δ371 single- and pu.1Δ371;spi-b∆232 double-mutant embryos. (K-L) WISH of lyz expression in 36-hpf pu.1Δ371 single- and pu.1Δ371;spi-b∆232 double-mutant embryos. (M) Quantification of mfap4+ macrophages and lyz+ neutrophils as shown in panels I-J and K-L, respectively. n.s., not significant, ***P < .001, Student t test; mfap4+pu.1Δ371 (mean/standard error [SE]/n) = 39.0/3.0/16, mfap4+dmut (mean/SE/n) = 22.9/2.2/18, lyz+pu.1Δ371 (mean/SE/n)=117.4/4.4/7, lyz+dmut (mean/SE/n) =121.6/4.2/8). Errors bars represent SE.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/129/4/10.1182_blood-2016-07-727651/4/m_blood727651f5.jpeg?Expires=1769080560&Signature=eWKHsioryC94jHmjaRbbjzO6WitojJcnkj9VxTx4IiDnslAVLHn76LzPmS~HvHdrUwATz-9nuF1D1I0vwsODRMBZXuTM6otQ8rzknEcK~~gd0lwpZ05XF-0-xYVsGlguLOhIO~kznLch7sNwgEMAZkz06fXA2b962DsXVCfz3FWy9JAR7wO5VZavudUQBeaa--YRWmRf9JI4gC9XtOx5XN4DdYn76-~Ce7Q6QI4Aa2bV5E28nvGEuYWwyDoMpmhtR35tG3Fu5fH6pvEUHzY0yFgUmfZSAW5Bi0l6vjZiUH46IkskiaeB-9f-qsVJUyh-WetNM7941C9M39lKwFviFA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal