In this issue of Blood, Zhang et al show the cooperation between P53 deletion and NRASG12D in the development of acute myeloid leukemia (AML). They also shed light on the megakaryocyte-erythroid progenitor (MEP) being the leukemia-initiating cell (LIC) targeted by P53 deletion and NRASG12D cooperation in mice.1
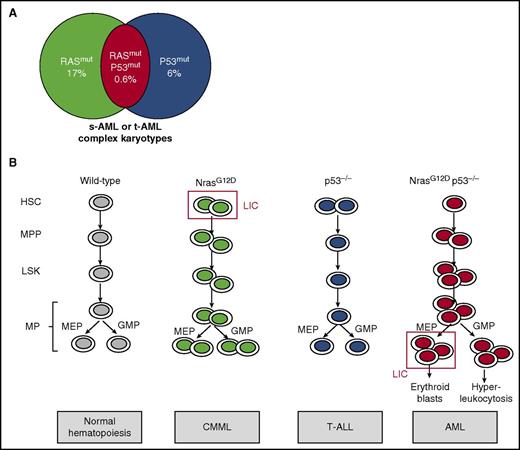
NrasG12D and p53 deletion cooperate to induce a full-blown erythromyeloid leukemia by MEP transformation. (A) Mutations in molecules of RAS pathway were found in 17% of AML cases, whereas P53 mutations were found in 6% of AML cases. Moreover, comutation was found in 0.6% of AML cases, which were secondary or therapy-related AML with complex cytogenetics. (B) NrasG12D KI mouse models presented an increase in hematopoietic stem cells (HSCs) and led to a CMML-like disease, whereas p53−/− mice have no major role in myeloid hematopoietic compartments and led to T-ALL. In contrast, NrasG12D p53−/− mice presented HSC, Lin–Sca1+c-Kit+ (LSK), multipotent progenitor (MPP), MEP, and granulocyte-monocyte progenitor (GMP) increases, leading to AML. The MEP was demonstrated to be the LIC. MP, myeloid progenitor; s-AML, secondary AML; t-AML, therapy-related AML.
NrasG12D and p53 deletion cooperate to induce a full-blown erythromyeloid leukemia by MEP transformation. (A) Mutations in molecules of RAS pathway were found in 17% of AML cases, whereas P53 mutations were found in 6% of AML cases. Moreover, comutation was found in 0.6% of AML cases, which were secondary or therapy-related AML with complex cytogenetics. (B) NrasG12D KI mouse models presented an increase in hematopoietic stem cells (HSCs) and led to a CMML-like disease, whereas p53−/− mice have no major role in myeloid hematopoietic compartments and led to T-ALL. In contrast, NrasG12D p53−/− mice presented HSC, Lin–Sca1+c-Kit+ (LSK), multipotent progenitor (MPP), MEP, and granulocyte-monocyte progenitor (GMP) increases, leading to AML. The MEP was demonstrated to be the LIC. MP, myeloid progenitor; s-AML, secondary AML; t-AML, therapy-related AML.
The RAS/MAPK signaling pathway mediates cell proliferation, and mutations in this axis have been frequently identified in patients harboring myeloid malignancies such as juvenile myelomonocytic leukemia or chronic myelomonocytic leukemia (CMML), and more rarely in myeloproliferative neoplasms (MPNs), myelodysplasia, and AML. They include somatic mutations in NRAS, KRAS, BRAF, NF1, and CBL, and SPRY4 deletion. These oncogenic events have been shown to be drivers of the disease in mice because interferon-induced Cre-mediated expression of oncogenic KrasG12D from its endogenous promoter in knock-in (KI) mice leads to a lethal CMML-like disease. Similarly, NrasG12D KI mice harbor a wide spectrum of myeloid diseases with a highly penetrant CMML in bone marrow transplantation assays.2,3 However, none of these models develop AML, suggesting that other associated genetic or epigenetic events are necessary to induce a full-blown leukemia.
TP53, or P53, is a tumor suppressor gene that encodes a transcription factor that plays a pleiotropic role in the cell cycle, DNA repair mechanism, apoptosis, autophagy, and senescence. Although P53 mutations have been frequently identified in solid tumors, they are present in <10% of AML. However, recent studies have shown that P53 mutations were associated with therapy-related AML with a very poor prognosis.4 Of note, frequent alterations in post-BCR-ABL-positive and -negative MPN leukemia involve P53 mutations (in ∼30% of cases) or P53 repression by negative regulators (MDM4 gene amplification [18% of cases] or posttranslational MDM2 upregulation).5,6 In the present study, Zhang et al identified P53 mutations, RAS mutations, or both mutations in 6%, 17%, and 0.6% of AML cases, respectively (see figure). Most cases with both mutations correspond to secondary- or therapy-related AML presenting complex karyotypes and of very poor prognosis compared with cases with RAS or P53 mutations only. Recent studies have shown that chemotherapy could select preexisting P53 clones, explaining the enrichment of P53 mutations in secondary leukemia.7 Moreover, complex cytogenetic studies indicate the presence of a high genetic instability, probably explained by the regulation of many DNA repair mechanisms by P53, such as a negative control of homologous recombination. Therefore, P53 alterations might favor persistent genetic and chromosomic abnormalities, a phenomenon that could be strengthened by chemotherapeutic agents.
In order to understand if P53 deletion could cooperate with RAS mutations to induce transformation into AML, Zhang et al used several mouse models. They showed that although transplantation of bone marrow cells with NrasG12D induced a CMML and with p53−/− an acute T-cell lymphoblastic leukemia/lymphoma (T-ALL), NrasG12Dp53−/− cell transplantation recapitulated an unexpected full-blown transplantable AML in 60% of animals (40% associated with T-ALL). These AML were characterized by a hyperleukocytosis and decreased hematocrit and platelets as well as an extramedullary accumulation of blasts cells in spleen and liver (see figure). Importantly, AML was also induced in primary NrasG12DP53−/− KI animals, thus without transplantation. These results are in line with previous results combining an activation of the RAS pathway and p53 silencing.8 A parallel could also be made with the overexpression of Jak2V617F in p53−/− mice, which induces leukemia.9
Zhang et al observed a major increase in MPPs and MPs in NrasG12Dp53−/− compared with controls. Importantly, these MPs displayed higher replating efficacy, and the transplantation of 10 000 MPs led exclusively to AML (as opposed to MPPs that led to both AML and T-ALL) in half of the animals, suggesting that they contain the LIC. By comparing the transcriptional profile of AML-MPs and control MPs, they found a specific upregulation of erythroid genes (Gata1/2, Epor, Fog1) together with enrichment of p53 target genes, pointing out the MEP as the potential LIC, although a downstream progenitor could also be the true LIC. These AML-MEPs were shown to both maintain an MEP signature and acquire an HSC signature. They were able to specifically induce AML after transplantation with only 2000 cells, whereas twice as many GMPs did not trigger any disease. Very interestingly, MEP was also the LIC in the Jak2V617F p53−/− AML model, suggesting that increased RAS signaling through JAK2V617F cooperating with P53 deletion was similarly involved in this leukemic process.
Furthermore, they demonstrated that NrasG12Dp53−/− mice present a very unexpected phenotype that associated increased proliferation and numbers of HSCs, LSK cells, and MPPs with an expansion of quiescent MEPs and GMPs. Finally, NrasG12D loss of heterozygosity was observed in some mice, showing that a higher RAS activity is required to induce AML in cooperation with P53 deletion. However, the clear mechanism of such cooperation remains incompletely understood. The AML presently described displays a defect in terminal erythroid differentiation combined with an upregulation of erythroid but not of megakaryocyte genes in MPs, suggesting that p53−/−NrasG12D promotes an erythroleukemia. This erythroid phenotype is in agreement with a report showing that MEP corresponds essentially to an immature cell committed to erythroid differentiation using single-cell RNA-Seq.10 There is previous evidence that the ERK pathway inhibits the terminal erythroid differentiation and that P53 may be important for proper erythroid differentiation. Thus, both gene alterations may cooperate to deregulate the erythroid differentiation program through transcription modification. Alternatively, they could also induce a higher genetic instability strengthened by chemotherapy promoting additional genetic alterations, both mechanisms being not mutually exclusive.
Finally, this work is of importance by showing that the MEP could be a common LIC for Ras activation and Jak2 activation in a p53-deleted context. It should be determined if similar cooperation could be seen with P53 mutations, which are not always a pure loss of function. This cooperation between p53 silencing and Nras activation raises the possibility of trying to inhibit the Ras pathway together with reactivating the P53 pathway in these patients.
Conflict-of-interest disclosure: The authors declare no competing financial interests.