In this issue of Blood, using induced pluripotent stem cell (iPSC)–derived human megakaryocytes (iMegs), Vo et al find that the megakaryocyte/platelet defects observed in patients with Paris-Trousseau syndrome (PTS) are due solely to hemizygous deletion of the transcription factor (TF) FLI1.1
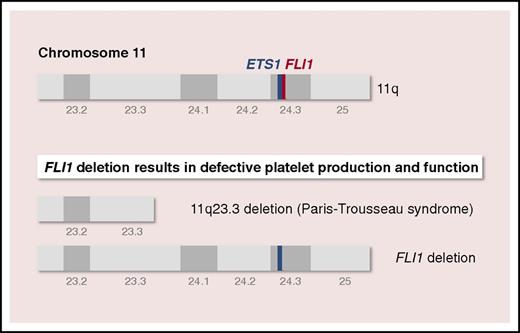
The genes encoding hematopoietic transcription factors ETS1 and FLI1 are located on chromosome 11q24.3, which is deleted in PTS. Using PTS-specific (11q23.3 deletion) and genome-edited (FLI1 deletion) induced pluripotent stem cell–derived iMegs, Vo et al find that the megakaryocyte/platelet defects observed in patients with PTS are due solely to hemizygous deletion of the TF FLI1.
The genes encoding hematopoietic transcription factors ETS1 and FLI1 are located on chromosome 11q24.3, which is deleted in PTS. Using PTS-specific (11q23.3 deletion) and genome-edited (FLI1 deletion) induced pluripotent stem cell–derived iMegs, Vo et al find that the megakaryocyte/platelet defects observed in patients with PTS are due solely to hemizygous deletion of the TF FLI1.
TFs bind to specific DNA sequences adjacent to the genes they regulate to promote or block the recruitment of RNA polymerase, thereby controlling the transcription of genetic information from DNA to RNA. Hematopoiesis critically depends on a complex network of TFs that temporally and spatially control the differentiation and maturation of individual lineages.2 TFs work alone or in complexes. For example, in megakaryopoiesis, the TFs GATA1, FOG1, and RUNX1 form a complex with TFs of the E26 transformation-specific (ETS) family that includes ETS1, GABPA, and FLI1 to regulate megakaryocyte-specific gene expression. Mutations in TFs have been discovered in hematopoietic malignancies, and there is now increasing evidence that these mutations are an important underlying cause for defects in platelet production, morphology, and function.3 These TFs include GATA1, RUNX1, FLI1, ETV6, and GFI1B.
Paris-Trousseau syndrome (Mendelian inheritance in man 188025) is a congenital platelet disorder encountered in most patients with Jacobsen syndrome (Mendelian inheritance in man 147791), a rare, inherited disorder characterized by skull dysmorphism, developmental delay, and multiple organ abnormalities. PTS is characterized by mild lifelong bleeding tendency, macrothrombocytopenia, bone marrow dysmegakaryopoiesis, and giant fused α-granules in a small population of platelets.4,5 Genetically, PTS is due to terminal deletion on the long arm of chromosome 11, at 11q23.3, a region that contains the genes encoding ETS1 and FLI1, both located on chromosome 11q24.3 (see figure). Whether PTS is due to hemizygous deletion of 1 or both genes is unclear.
Based on the study of primary CD34+ cells from patients with PTS, Raslova et al concluded in 2004 that the platelet disorder is due to allelic exclusion of FLI1 in CD41+/CD42– progenitors, in which only the subpopulation of megakaryocytes originating from progenitors missing FLI1 are affected.6 More recently, missense mutations and deletions in the DNA-binding domain of FLI1 have been implicated as the probable cause of the megakaryocyte/platelet defects observed in patients with PTS.7,8 Although these reports suggest that FLI1 hemizygous deletion may be an important cause of the inherited thrombocytopenia, they did not detail the platelet defect or compare them with PTS platelets. Mice lacking FLI1 die of embryonic hemorrhage that is attributed to the role of FLI1 in both megakaryopoiesis and endothelium and hemangioblast specification.9,10 However, hemizygous FLI1 deletion in mice does not recapitulate the human PTS phenotype.
To determine whether the megakaryocyte/platelet defects observed in patients with PTS were specifically the result of FLI1 hemizygous deletion and not to that of ETS1, Vo et al generated PTS-specific (11q23.3 deletion) and genome-edited (FLI1 deletion) iPSC-derived iMegs and platelets (see figure). The study compares iMegs derived from a patient with PTS and iMegs in which FLI1 is hemizygously deleted (FLI1+/−) using transcription activator-like effector nucleases. PTS and FLI1+/− iMegs replicate many of the described megakaryocyte/platelet features of PTS, including a decrease in iMeg yield and fewer platelets released per iMeg. Further, platelets from these FLI1-deficient human iMegs released in nonobese diabetic/severe combined immunodeficiency/interleukin-2 receptor γ-chain deficient (NSG) mice have poor half-lives and functionality, confirming the central role of FLI1 hemizygous deletion in the megakaryocyte and platelet defects observed in PTS.
Importantly, ETS1 is overexpressed in PTS and FLI1+/− iMegs, suggesting that FLI1 negatively regulates ETS1 in megakaryopoiesis and that the megakaryocyte/platelet defects observed in patients with PTS are unlikely to involve ETS1 allelic exclusion or deficiency, and are due solely to hemizygous deletion of FLI1.
Vo et al also overexpressed FLI1 in control and PTS iMegs using the glycoprotein Ibα promoter for megakaryocyte-specific expression and observed that FLI1 overexpression increases the yield of control iMegs in vitro and the yield, half-life, and functionality of released platelets in NSG mice, demonstrating that FLI1 overexpression enhances megakaryopoiesis, platelet production, and function.
In conclusion, the study by Vo et al confirms the central role of FLI1 hemizygous deletion in the megakaryocyte and platelet defects observed in PTS and demonstrates that FLI1 overexpression enhances megakaryopoiesis, thrombopoiesis, and platelet biology. Many questions remain unanswered; for example, whether FLI1 deletion affects the expression of other hematopoietic TFs such as GATA1, and whether ETS1 deletion reciprocally affects FLI1 expression remain to be addressed. Further, whether PTS and FLI1+/− iMegs exist as 2 subpopulations, because of allelic FLI1exclusion, as described by Raslova et al for CD34+ derived PTS megakaryocytes,6 is unclear.
Conflict-of-interest disclosure: The author declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal